- The paper presents TrialMind, an AI framework that automates study search, screening, data extraction, and evidence synthesis for systematic clinical reviews.
- It employs large language models with prompt engineering and retrieval-augmented generation to achieve high recall (0.897-1.000) in literature search and accurate data extraction.
- Human evaluators preferred TrialMind over traditional methods, highlighting its potential to enhance the timeliness and reliability of clinical evidence synthesis.
Accelerating Clinical Evidence Synthesis with LLMs
The paper "Accelerating Clinical Evidence Synthesis with LLMs" introduces TrialMind, a generative AI-based framework designed to streamline the process of conducting medical systematic reviews. By employing LLMs, TrialMind offers a sophisticated pipeline that automates paper search, screening, data extraction, and clinical evidence synthesis, while integrating human oversight to ensure accuracy and reliability.
Introduction and Motivation
Systematic reviews are crucial for consolidating evidence from clinical studies, yet traditional methods are labor-intensive and time-consuming, often taking years to complete. The authors acknowledge the potential of LLMs to revolutionize this process by facilitating efficient and comprehensive literature reviews. TrialMind represents an advancement in applying LLMs to orchestrate the systematic review process through a structured and transparent pipeline, enhancing the timeliness and quality of clinical evidence synthesis.
TrialMind Framework
TrialMind comprises four key components: paper search, screening, data extraction, and evidence synthesis. Each component is driven by LLMs, which are enhanced by prompt engineering and retrieval-augmented generation techniques. These components are depicted in the TrialMind framework (Figure 1).
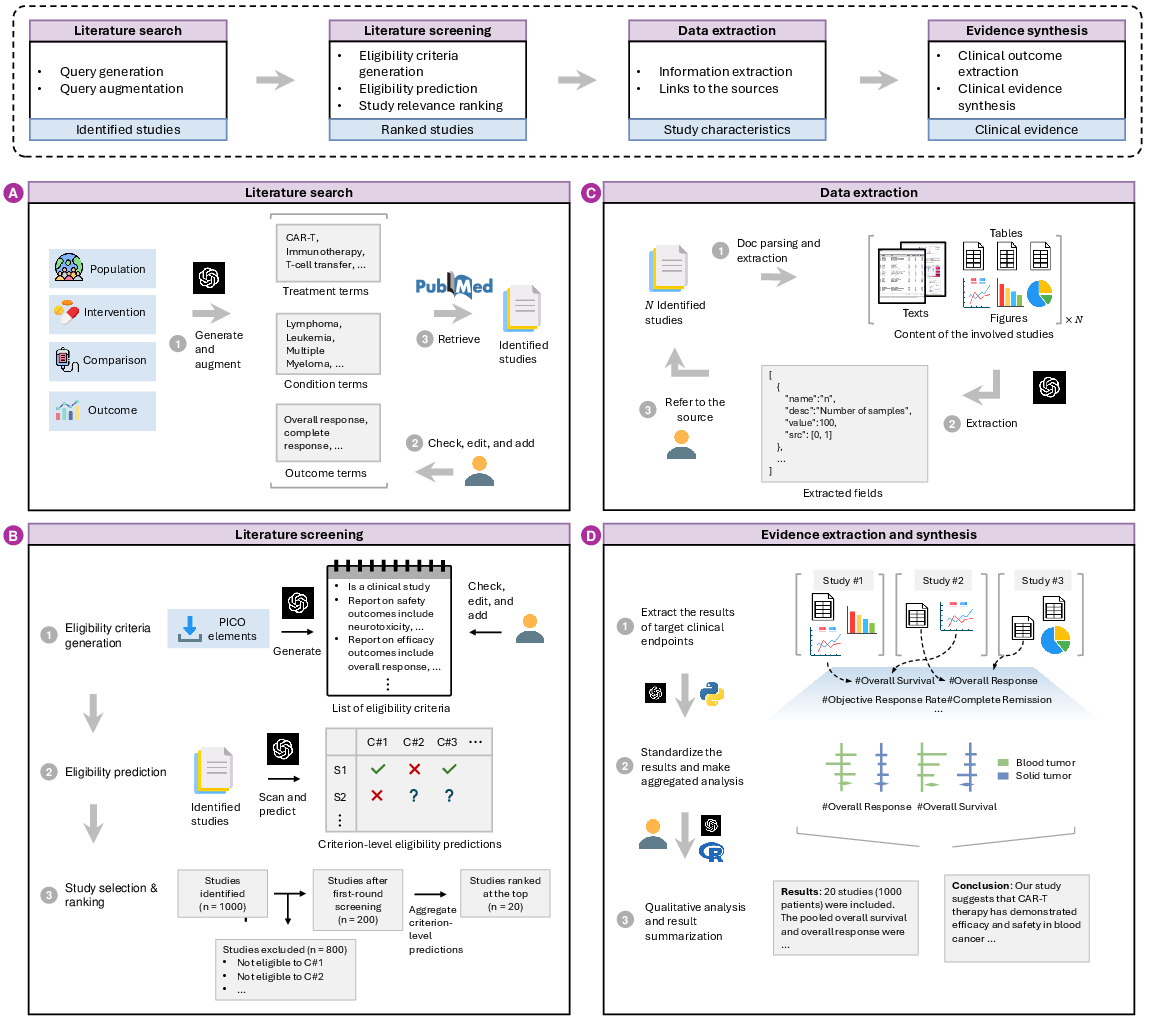
Figure 1: The TrialMind framework has four components: a) Utilizing input PICO elements, TrialMind generates key terms to construct Boolean queries for retrieving studies from literature databases. b) TrialMind formulates eligibility criteria, which users can edit to provide context for LLMs during eligibility predictions. Users can then select studies based on these predictions and rank their relevance by aggregating them. c) TrialMind processes the descriptions of target data fields to extract and output the required information as structured data. d) TrialMind extracts findings from the studies and collaborates with users to synthesize the clinical evidence.
Literature Search and Screening
TrialMind employs LLMs to generate Boolean queries from the PICO (Population, Intervention, Comparison, Outcome) elements of the research question, achieving high recall (0.897-1.000) in retrieving studies from vast databases such as PubMed. The approach integrates query generation, augmentation, and refinement to capture a comprehensive set of relevant studies (Figure 2).
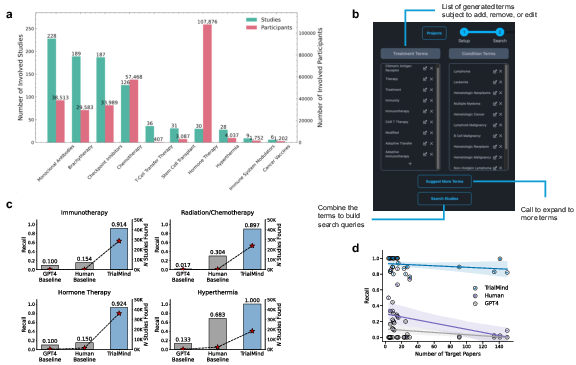
Figure 2: Literature search experiment results. a, Total number of involved studies and participants of the review papers across different topics. b, The TrialMind's interface for users to retrieve studies. c, The Recall of the search results for reviews across four topics. The bar heights indicate the Recall, and the star indicates the number of studies found. d, Scatter plots of the Recall against the number of ground-truth studies. Each scatter indicates the results of one review. Regression estimates are displayed with the 95\% CIs in blue or purple.
TrialMind uses LLMs to extract structured data from unstructured documents, integrating context-awareness to improve accuracy in extracting information such as paper characteristics, population baselines, and findings (Figure 3). This component surpasses baseline performance by providing a solid initial data extraction, which human experts can further refine.

Figure 3: Data and result extraction experiment results. a, Streamline paper information extraction using TrialMind. b, Data extraction accuracy within each field type across four topics. c, Confusion matrix showing the hallucination and missing rates in the data extraction results. d, Result extraction accuracy across topics. e, Result extraction accuracy across clinical endpoints. f, Error analysis of the result extraction. g, Streamline result extraction using TrialMind.
Evidence Synthesis
TrialMind synthesizes clinical evidence into formats suitable for meta-analysis, such as forest plots. Human evaluators favored TrialMind over traditional GPT-4 applications for its effectiveness and reliability, achieving a winning rate against the baseline across reviewed studies (Figure 4).
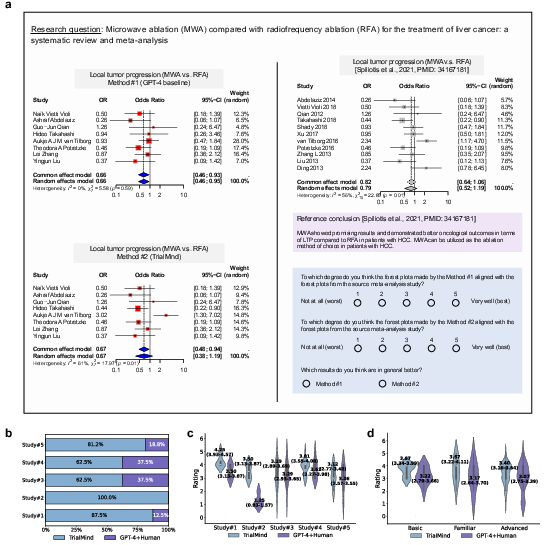
Figure 4: Human evaluation of the synthesized clinical evidence. a, The paper design compares the synthesized clinical evidence from the baseline and TrialMind via human evaluation. b, Winning rate of TrialMind against the GPT-4+Human baseline across studies. c, Violin plots of the ratings across studies. Each plot is tagged with the mean ratings (95\% CI) from all the annotators. d,Violin plots of the ratings across annotators with different expertise levels. Each plot is tagged with the mean ratings (95\% CI) from all the studies.
Implications and Future Work
TrialMind exemplifies the capability of LLMs to augment human expertise in medical systematic reviews. However, it emphasizes the necessity of human oversight to ensure error correction and contextual awareness throughout the pipeline. Future research could expand TrialMind to other areas of medicine and fine-tune prompt designs to further enhance effectiveness, providing more efficient and trustworthy methods in clinical research.
Conclusion
This paper introduces TrialMind as a transformative tool for clinical evidence synthesis, demonstrating its utility in automating library searches, screening, data extraction, and evidence synthesis with remarkable accuracy and reliability. The integration of generative AI with rigorous human oversight represents a scalable solution to the challenges faced in medical systematic reviews, paving the way for more timely and reliable medical advancements.





