Tx-LLM: A Large Language Model for Therapeutics (2406.06316v1)
Abstract: Developing therapeutics is a lengthy and expensive process that requires the satisfaction of many different criteria, and AI models capable of expediting the process would be invaluable. However, the majority of current AI approaches address only a narrowly defined set of tasks, often circumscribed within a particular domain. To bridge this gap, we introduce Tx-LLM, a generalist LLM fine-tuned from PaLM-2 which encodes knowledge about diverse therapeutic modalities. Tx-LLM is trained using a collection of 709 datasets that target 66 tasks spanning various stages of the drug discovery pipeline. Using a single set of weights, Tx-LLM simultaneously processes a wide variety of chemical or biological entities(small molecules, proteins, nucleic acids, cell lines, diseases) interleaved with free-text, allowing it to predict a broad range of associated properties, achieving competitive with state-of-the-art (SOTA) performance on 43 out of 66 tasks and exceeding SOTA on 22. Among these, Tx-LLM is particularly powerful and exceeds best-in-class performance on average for tasks combining molecular SMILES representations with text such as cell line names or disease names, likely due to context learned during pretraining. We observe evidence of positive transfer between tasks with diverse drug types (e.g.,tasks involving small molecules and tasks involving proteins), and we study the impact of model size, domain finetuning, and prompting strategies on performance. We believe Tx-LLM represents an important step towards LLMs encoding biochemical knowledge and could have a future role as an end-to-end tool across the drug discovery development pipeline.
Sponsor
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
Plain-language summary of “Tx-LLM: A LLM for Therapeutics”
What is this paper about?
The paper introduces Tx-LLM, a large AI model designed to help with drug discovery. Instead of building lots of small, separate tools for each step in finding and testing new medicines, Tx-LLM is one general model that can handle many different tasks using the same set of learned skills.
What questions did the researchers ask?
In simple terms, they wanted to know:
- Can one big AI model learn enough about chemistry and biology to help with many steps in drug development?
- Will training it on many kinds of data (like drug molecules, protein sequences, and disease names) make it better at each individual task?
- How do things like model size, the way it’s trained, and how we ask it questions affect its performance?
How did they try to answer these questions?
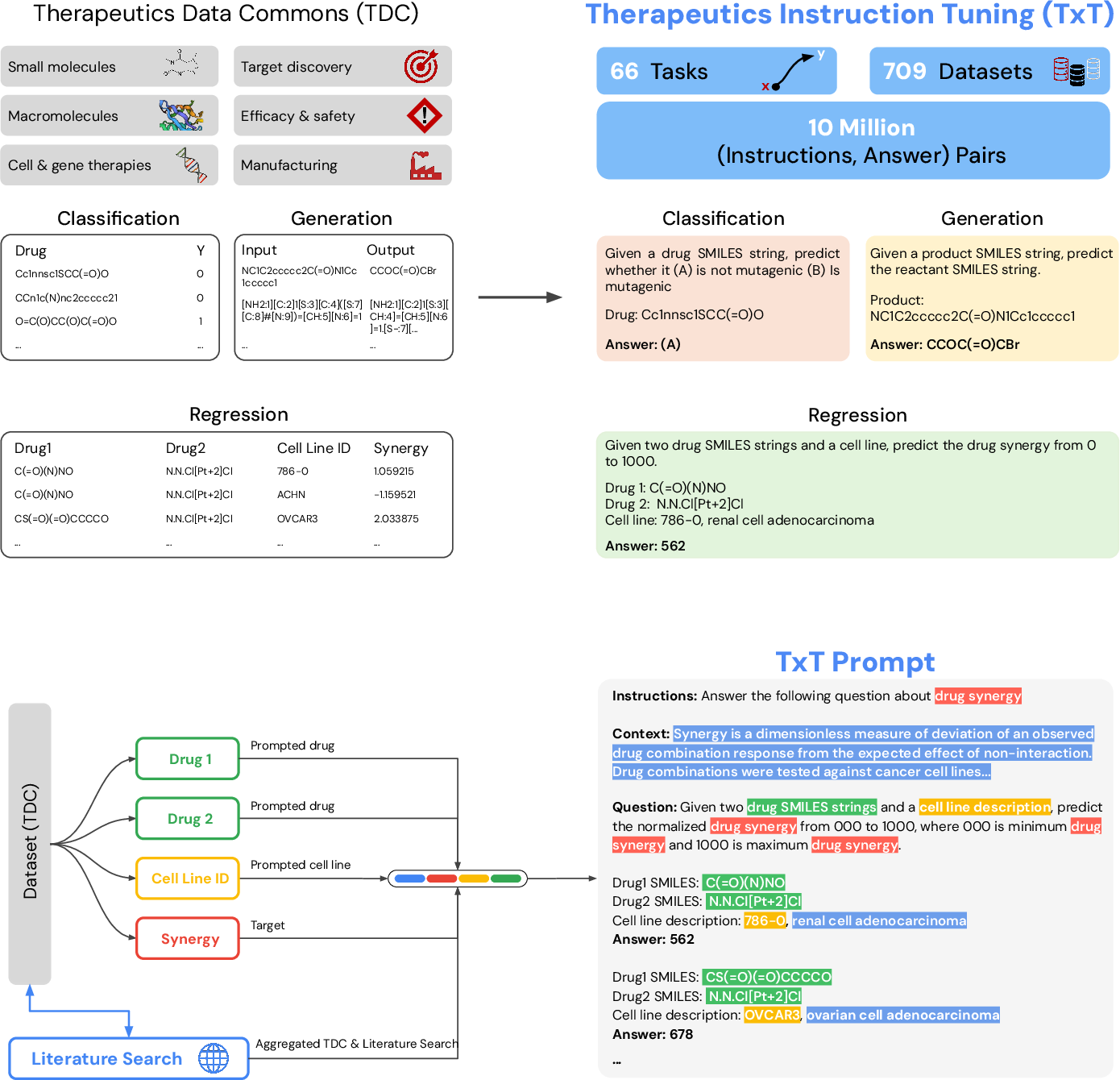
They built a training collection called TxT using 709 datasets that cover 66 different tasks from across the drug discovery pipeline (for example, predicting whether a drug is toxic, whether it binds to a target, how well it might work, and even whether a clinical trial could succeed).
To make this understandable to a LLM, they turned all inputs into text:
- Small molecules were written as SMILES strings, which are like special “codes” that describe a molecule.
- Proteins and genes were written as their letter sequences (amino acids or nucleotides).
- Extra details (like disease names or cell line descriptions) stayed as regular text.
They then fine-tuned an existing LLM (PaLM‑2) so it could read prompts that look a bit like short quiz questions. Each prompt had:
- An instruction (what to do),
- A short context (why this matters),
- A question (the task itself, mixing text with molecule or protein strings),
- And an answer (for example, yes/no, a number, or generated text).
Some tasks were:
- Classification (yes/no, like “Is this drug toxic?”),
- Regression (a number, like “How strong is the binding?”; they turned numbers into “bins” from 000 to 1000 so the model could predict a label),
- Generation (like predicting reactants from a product in a chemical reaction).
They tested different ways of asking the model questions (with zero examples or a few examples included in the prompt), compared different model sizes, and checked how much domain-specific training helped.
What did they discover?
Here are the main results:
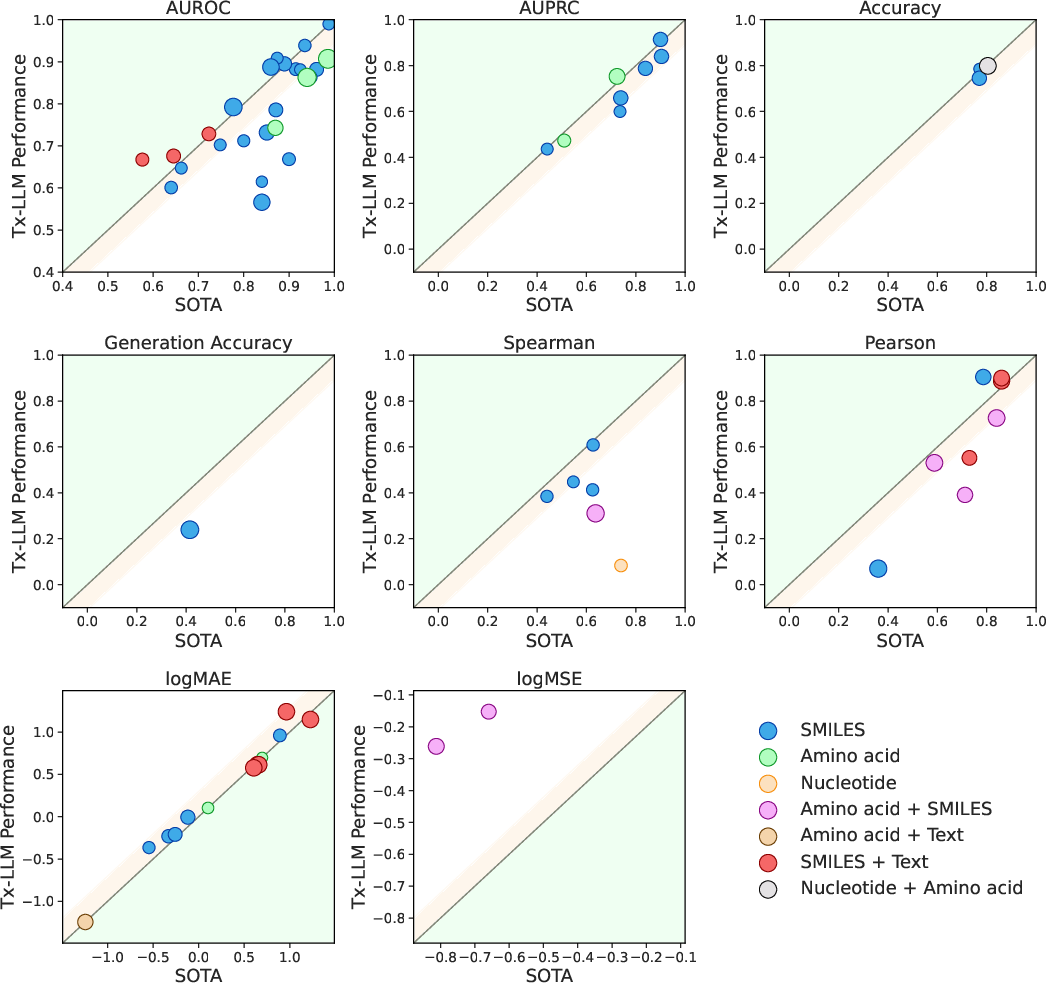
- Strong overall performance: Out of 66 tasks, Tx-LLM matched or beat the best published results on 43. It set new best results on 22 of them, all while using one single model for everything.
- Best when mixing molecules with text: Tasks that combined a molecule (SMILES) plus text (like disease or cell line names) worked especially well. This likely benefits from the model’s general language knowledge learned before fine-tuning.
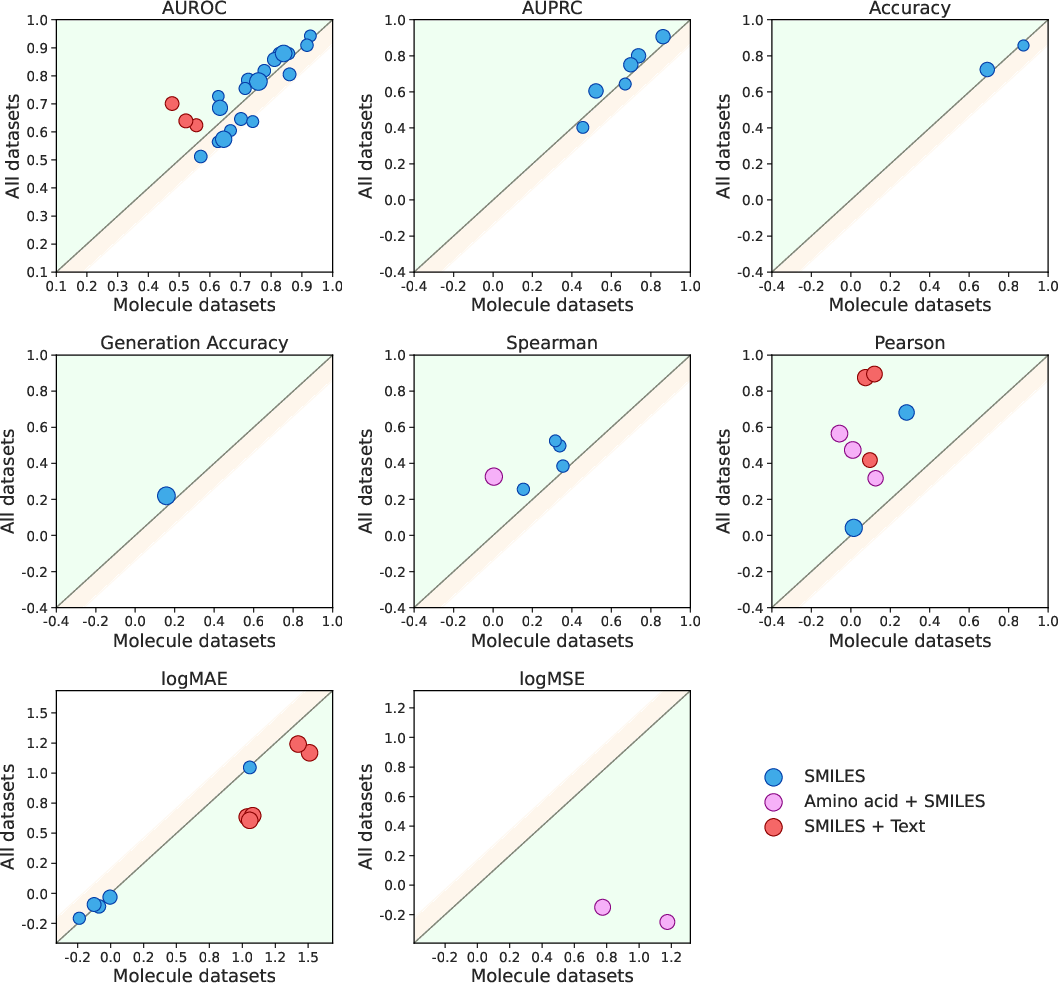
- Learning transfers across types: Training on very different drug types (like proteins and small molecules) helped the model do better overall. In other words, learning about protein sequences also improved predictions about small molecules—this is called positive transfer.
- Bigger and more specialized training helps: Larger model versions did better, and fine-tuning on therapeutic data clearly improved results compared to the base (unadapted) model.
- Context matters: Adding helpful background sentences in the prompt improved performance for most tasks.
- Few-shot vs zero-shot: Adding a few examples to the prompt didn’t consistently beat zero examples; both worked similarly overall.
- Limits: On tasks that use only SMILES strings (no extra text), some specialist models that represent molecules as graphs still did better. Also, LLMs can sometimes write invalid SMILES strings, which is like making up impossible molecules.
Why is this important?
If one AI model can handle many different steps in drug discovery, it could help scientists move faster and spend less money by:
- Screening out weak or risky drug candidates earlier,
- Prioritizing which experiments to run in the lab,
- Linking together knowledge about chemistry, biology, and diseases in one place.
That said, Tx-LLM is still research. It doesn’t replace real experiments, and it isn’t perfect on every task. It also wasn’t tuned to explain its answers in natural language, and care is needed to avoid mistakes like invalid molecule strings. Still, this work is a promising step toward an “all-in-one” AI assistant that supports many stages of medicine development—from picking targets to predicting clinical trial success.
Collections
Sign up for free to add this paper to one or more collections.