RegWSI: Whole Slide Image Registration using Combined Deep Feature- and Intensity-Based Methods: Winner of the ACROBAT 2023 Challenge (2404.13108v2)
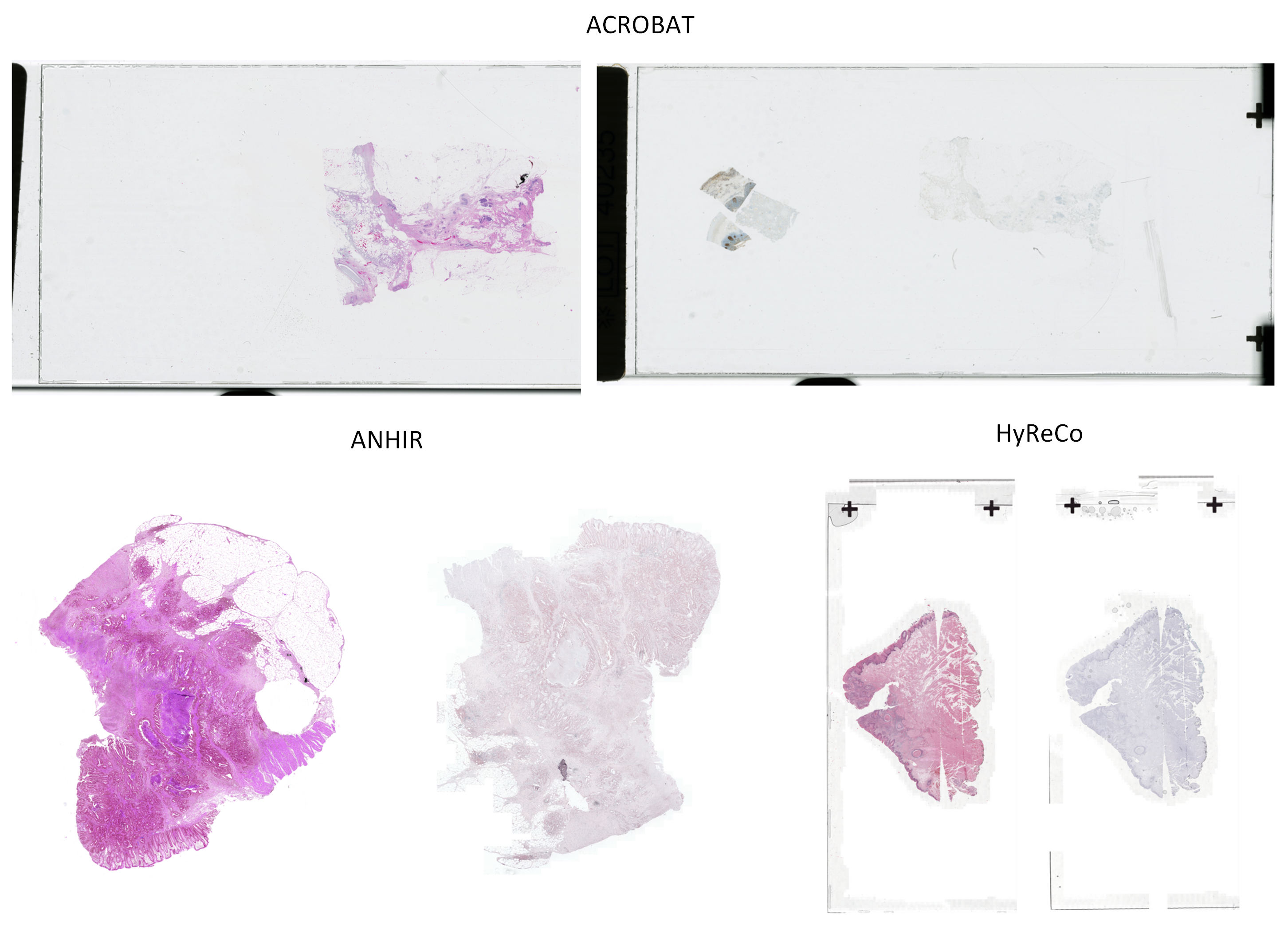
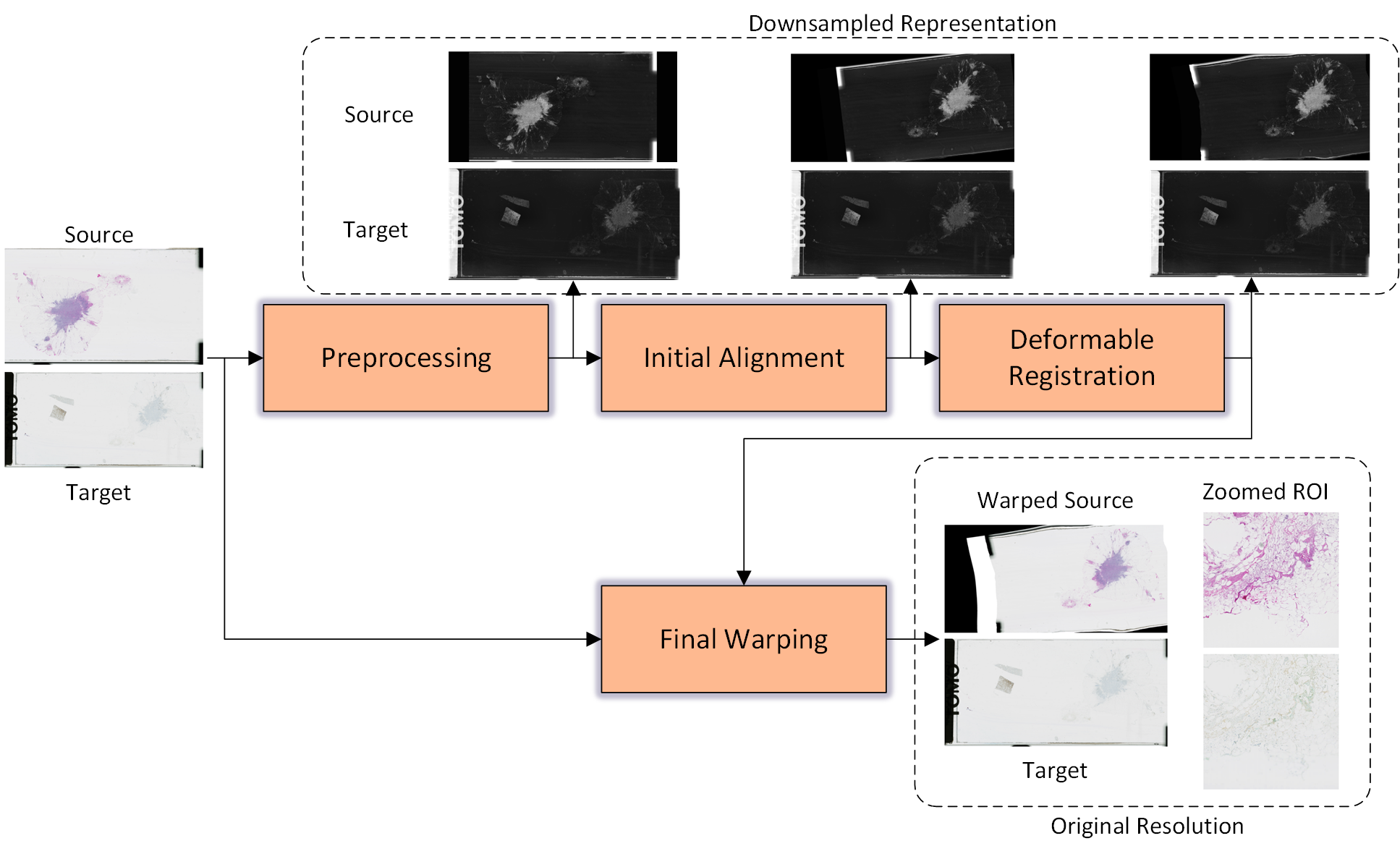
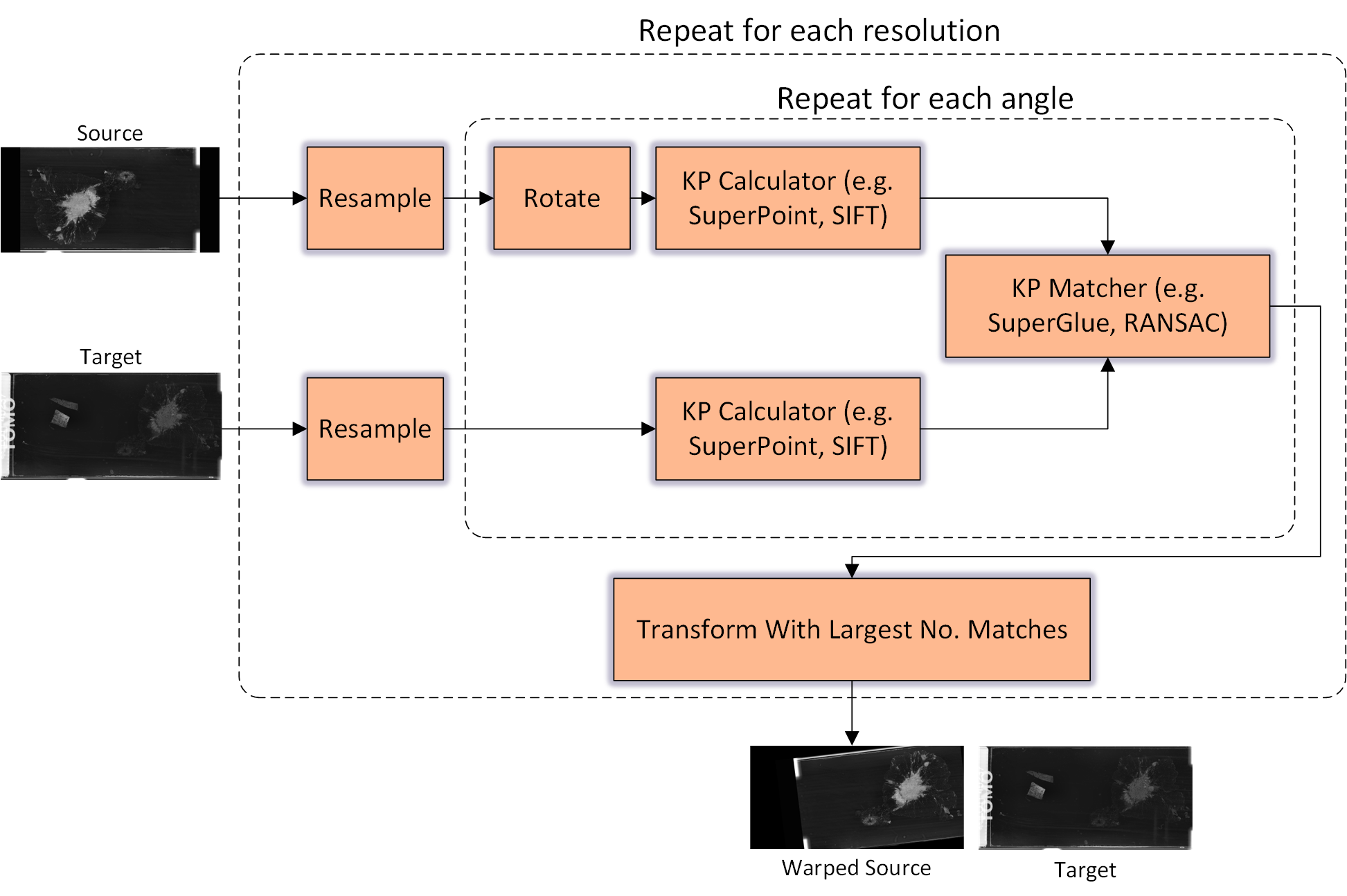
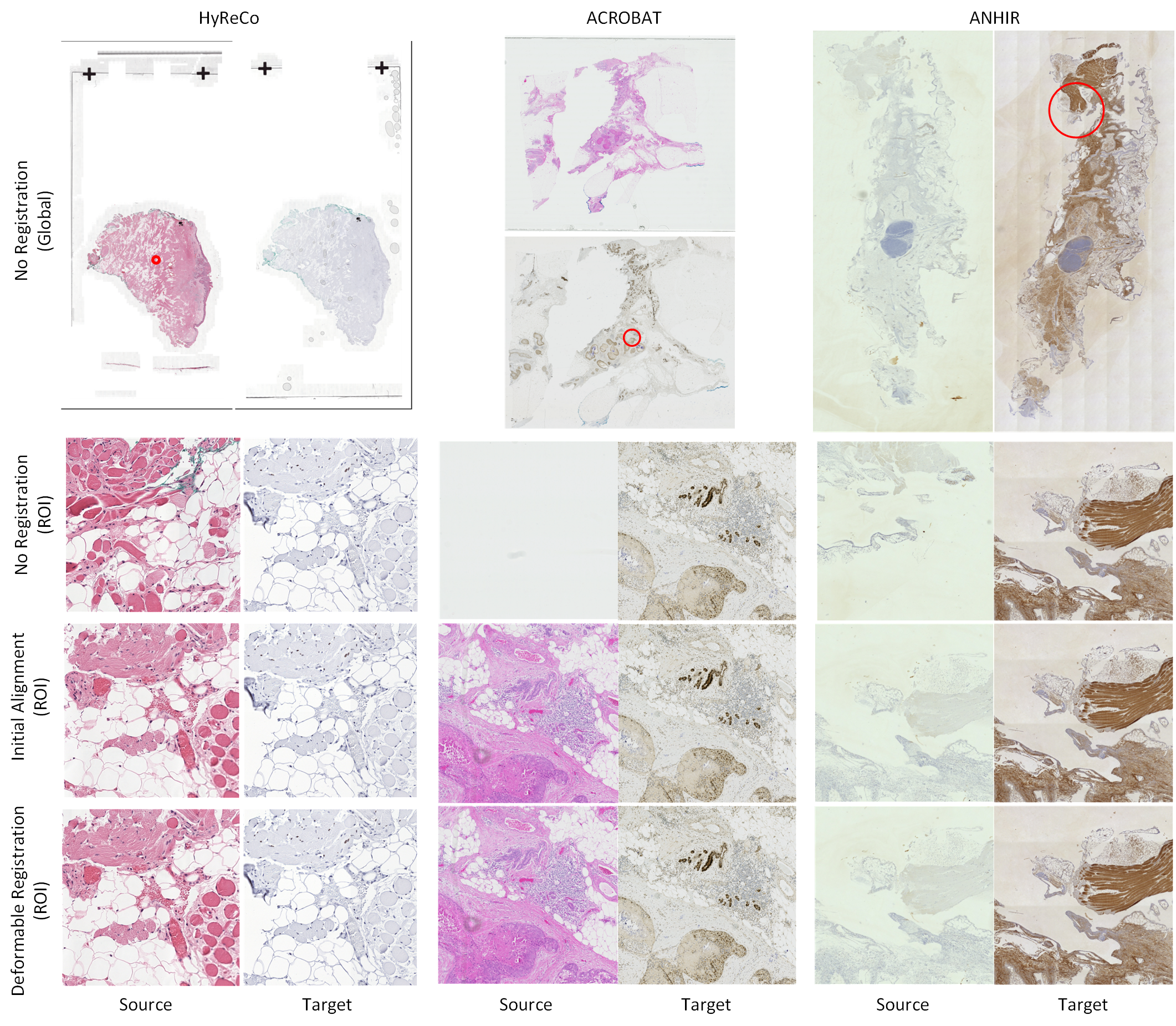
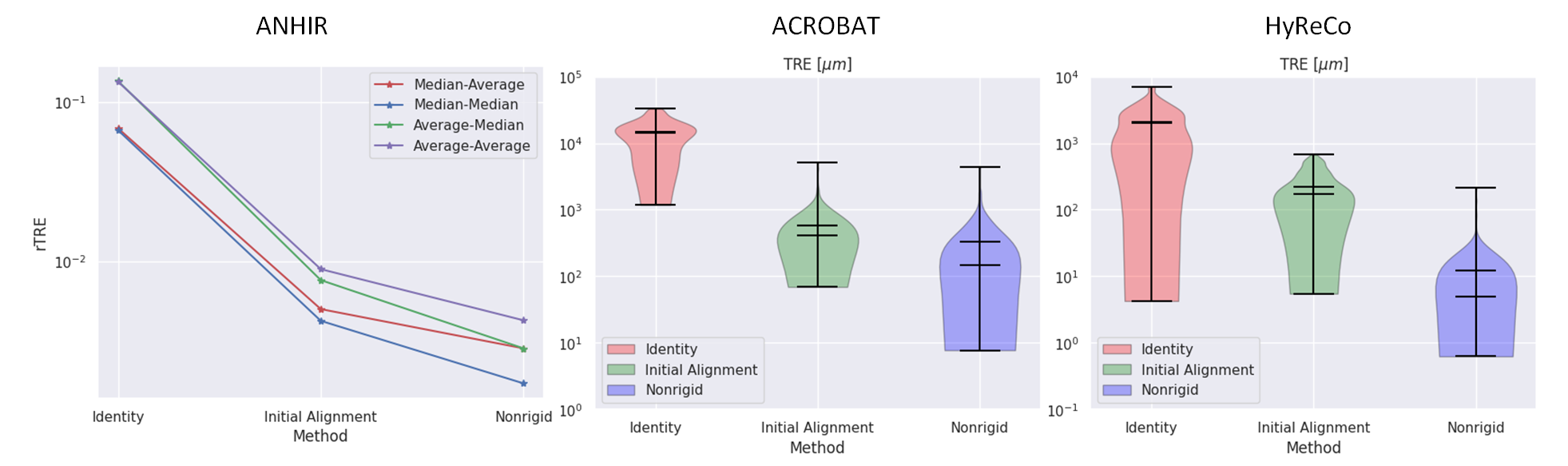
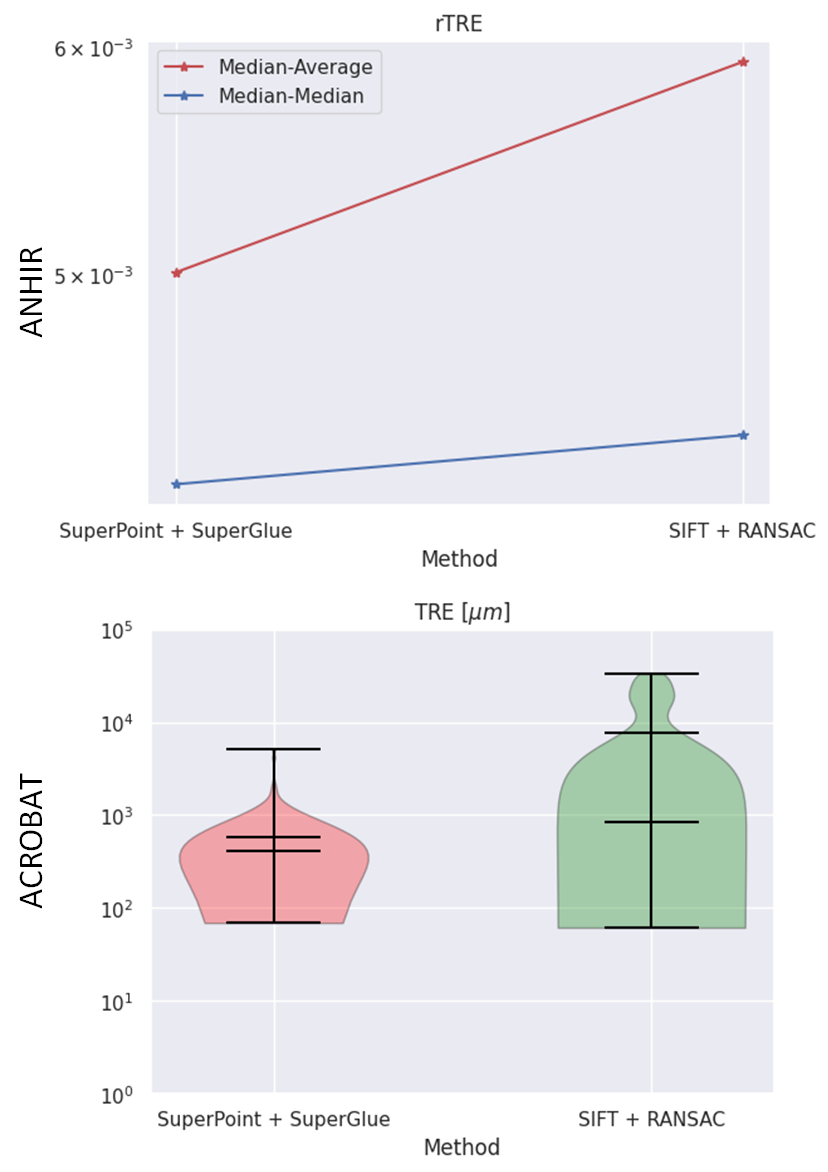
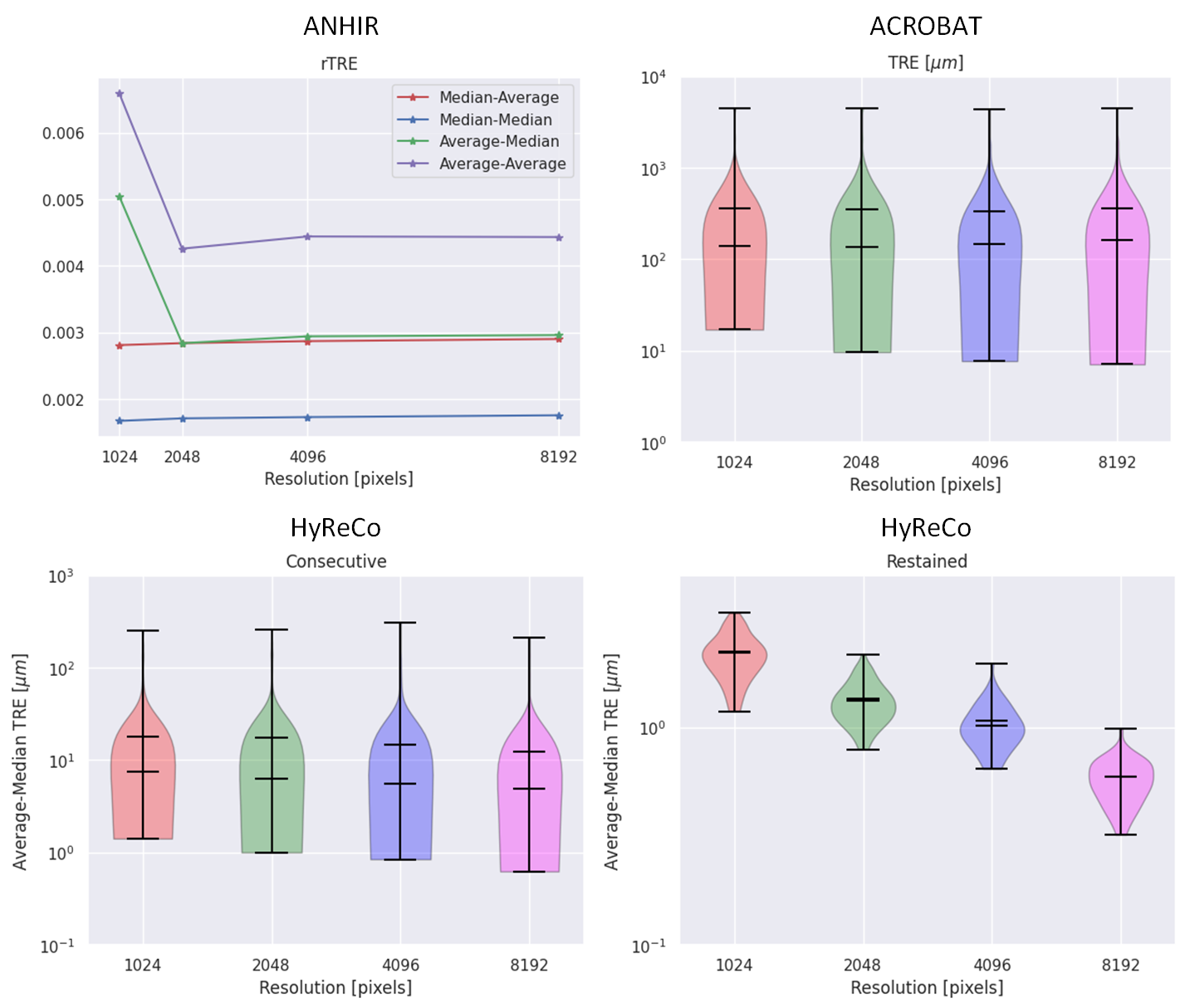
Abstract: The automatic registration of differently stained whole slide images (WSIs) is crucial for improving diagnosis and prognosis by fusing complementary information emerging from different visible structures. It is also useful to quickly transfer annotations between consecutive or restained slides, thus significantly reducing the annotation time and associated costs. Nevertheless, the slide preparation is different for each stain and the tissue undergoes complex and large deformations. Therefore, a robust, efficient, and accurate registration method is highly desired by the scientific community and hospitals specializing in digital pathology. We propose a two-step hybrid method consisting of (i) deep learning- and feature-based initial alignment algorithm, and (ii) intensity-based nonrigid registration using the instance optimization. The proposed method does not require any fine-tuning to a particular dataset and can be used directly for any desired tissue type and stain. The method scored 1st place in the ACROBAT 2023 challenge. We evaluated using three open datasets: (i) ANHIR, (ii) ACROBAT, and (iii) HyReCo, and performed several ablation studies concerning the resolution used for registration and the initial alignment robustness and stability. The method achieves the most accurate results for the ACROBAT dataset, the cell-level registration accuracy for the restained slides from the HyReCo dataset, and is among the best methods evaluated on the ANHIR dataset. The method does not require any fine-tuning to a new datasets and can be used out-of-the-box for other types of microscopic images. The method is incorporated into the DeeperHistReg framework, allowing others to directly use it to register, transform, and save the WSIs at any desired pyramid level. The proposed method is a significant contribution to the WSI registration, thus advancing the field of digital pathology.
- ANHIR: automatic non-rigid histological image registration challenge. IEEE transactions on medical imaging, 39(10):3042–3052, 2020.
- Increasing the usefulness of already existing annotations through wsi registration. arXiv preprint arXiv:2303.06727, 2023a.
- 3d reconstruction of multiple stained histology images. Journal of pathology informatics, 4(2):7, 2013a.
- A review of artifacts in histopathology. Journal of oral and maxillofacial pathology: JOMFP, 22(2):279, 2018.
- HistoQC: an open-source quality control tool for digital pathology slides. JCO clinical cancer informatics, 3:1–7, 2019.
- M. Wodzinski. DeeperHistReg Library. https://github.com/MWod/DeeperHistReg/, 2023. [Online; accessed 14-December-2023].
- DeeperHistReg: Robust Whole Slide Images Registration Framework. In Review, 2024.
- The ACROBAT 2022 Challenge: Automatic Registration Of Breast Cancer Tissue. arXiv preprint arXiv:2305.18033, 2023b.
- ACROBAT - a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology. arXiv preprint arXiv:2211.13621, 2022.
- Comparison of consecutive and restained sections for image registration in histopathology. Journal of Medical Imaging, 2023.
- HyReCo - Hybrid Re-Stained And Consecutive Histological Serial Sections. IEEE Data Port, 2021.
- End-to-end affine registration framework for histopathological images with weak annotations. Computer Methods and Programs in Biomedicine, 241:107763, 2023.
- Deep feature based cross-slide registration. Computerized Medical Imaging and Graphics, 104:102162, 2023.
- Deformable medical image registration: A survey. IEEE transactions on medical imaging, 32(7):1153–1190, 2013.
- Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. IEEE Transactions on Medical Imaging, 42(3):697–712, 2022.
- Large deformation diffeomorphic image registration with laplacian pyramid networks. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part III 23, pages 211–221. Springer, 2020.
- Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging, 38(8):1788–1800, 2019.
- Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging, 29(1):196–205, 2009.
- Deep learning in medical image registration: a survey. Machine Vision and Applications, 31(1):8, 2020.
- Deformable medical image registration using generative adversarial networks. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pages 1449–1453. IEEE, 2018.
- Simpleelastix: A user-friendly, multi-lingual library for medical image registration. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pages 134–142, 2016.
- Deformable medical image registration: setting the state of the art with discrete methods. Annual review of biomedical engineering, 13:219–244, 2011.
- Mind: Modality independent neighbourhood descriptor for multi-modal deformable registration. Medical image analysis, 16(7):1423–1435, 2012.
- Medical image registration using mutual information. Proceedings of the IEEE, 91(10):1699–1722, 2003.
- Benchmarking of image registration methods for differently stained histological slides. In 2018 25th IEEE International Conference on Image Processing (ICIP), pages 3368–3372. IEEE, 2018.
- Robust, fast and accurate: a 3-step method for automatic histological image registration. arXiv preprint arXiv:1903.12063, 2019.
- Virtual alignment of pathology image series for multi-gigapixel whole slide images. Nature communications, 14(1):4502, 2023.
- Accurate and robust alignment of variable-stained histologic images using a general-purpose greedy diffeomorphic registration tool. arXiv preprint arXiv:1904.11929, 2019.
- Accurate and robust alignment of differently stained histologic images based on greedy diffeomorphic registration. Applied Sciences, 11(4):1892, 2021.
- Multistep, automatic and nonrigid image registration method for histology samples acquired using multiple stains. Physics in Medicine & Biology, 66(2):025006, 2021.
- Unsupervised content classification based nonrigid registration of differently stained histology images. IEEE Transactions on Biomedical Engineering, 61(1):96–108, 2013b.
- Patch-based nonlinear image registration for gigapixel whole slide images. IEEE Transactions on Biomedical Engineering, 63(9):1812–1819, 2015.
- Robust quad-tree based registration on whole slide images. In MICCAI Workshop on Computational Pathology, pages 181–190. PMLR, 2021.
- Unsupervised 3d end-to-end medical image registration with volume tweening network. IEEE journal of biomedical and health informatics, 24(5):1394–1404, 2019.
- DeepHistReg: Unsupervised deep learning registration framework for differently stained histology samples. Computer methods and programs in biomedicine, 198:105799, 2021.
- Unsupervised learning-based nonrigid registration of high resolution histology images. In Machine Learning in Medical Imaging: 11th International Workshop, MLMI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4, 2020, Proceedings 11, pages 484–493. Springer, 2020a.
- Unsupervised histological image registration using structural feature guided convolutional neural network. IEEE Transactions on Medical Imaging, 41(9):2414–2431, 2022.
- VA Pyatov and DV Sorokin. Affine registration of histological images using transformer-based feature matching. Pattern Recognition and Image Analysis, 32(3):626–630, 2022a.
- Tahir: Transformer-based affine histological image registration. In International Conference on Pattern Recognition, pages 541–552. Springer, 2022b.
- Learning-based affine registration of histological images. In Biomedical Image Registration: 9th International Workshop, WBIR 2020, Portorož, Slovenia, December 1–2, 2020, Proceedings 9, pages 12–22. Springer, 2020b.
- PyVips. PyVips Library. https://www.libvips.org/, 2023. [Online; accessed 14-December-2023].
- Superpoint: Self-supervised interest point detection and description. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pages 224–236, 2018.
- Superglue: Learning feature matching with graph neural networks. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pages 4938–4947, 2020.
- ANHIR Evaluation Platform. https://anhir.grand-challenge.org/evaluation/challenge/leaderboard/. [Online; accessed 14-December-2023].
- R. Fernandez-Gonzalez et al. System for combined three-dimensional morphological and molecular analysis of thick tissue specimens. Microscopy Research and Technique, 59(6):522–530, 2002.
- L. Gupta et al. Stain independent segmentation of whole slide images: A case study in renal histology. Proceedings - International Symposium on Biomedical Imaging, pages 1360–1364, 2018.
- The immune microenvironment of various histological types of ebv-associated gastric cancer. Virchows Archiv, 2018.
- G. Bueno and O. Deniz. AIDPATH: Academia and Industry Collaboration for Digital Pathology. http://aidpath.eu.
- ACROBAT Evaluation Platform. https://acrobat.grand-challenge.org/evaluation/model-development/leaderboard/. [Online; accessed 14-December-2023].
- Resolution-agnostic tissue segmentation in whole-slide histopathology images with convolutional neural networks. PeerJ, 7:e8242, 2019.
- Robust Multiresolution and Multistain Background Segmentation in Whole Slide Images. In Polish Conference on Biocybernetics and Biomedical Engineering, pages 29–40. Springer, 2023.
- BigPicture project. https://bigpicture.eu//. [Online; accessed 14-December-2023].
- Deformable Medical Image Registration Under Distribution Shifts with Neural Instance Optimization. In International Workshop on Machine Learning in Medical Imaging, pages 126–136. Springer, 2023.
Sponsor
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Collections
Sign up for free to add this paper to one or more collections.