- The paper presents the MedCLIP-SAM framework that integrates CLIP and SAM models for text-guided segmentation in both zero-shot and weakly supervised settings.
- It introduces the unique DHN-NCE loss to optimize BiomedCLIP fine-tuning, improving performance on sparse, small-batch medical datasets.
- Experimental results across breast ultrasound, brain MRI, and lung X-ray modalities show superior metrics like IoU, DSC, and AUC compared to baselines.
Introduction
Medical image segmentation is a critical aspect of modern clinical practices, aiding in diagnosis, study, and treatment planning. Traditional deep learning-based segmentation models have advanced the field, yet struggle with issues of data efficiency, generalizability, and interactivity. Addressing these challenges, foundation models such as CLIP and SAM offer promising cross-domain representational capabilities, though their application to medical imaging remains underexplored.
MedCLIP-SAM is an innovative framework that combines CLIP and SAM models to facilitate medical image segmentation using text prompts, operational in both zero-shot and weakly supervised paradigms. This framework introduces the Decoupled Hard Negative Noise Contrastive Estimation (DHN-NCE) loss, enhancing the fine-tuning of BiomedCLIP. Additionally, it leverages gScoreCAM to formulate prompts, thereby generating segmentation masks in a zero-shot context, which can be refined in weak supervision.
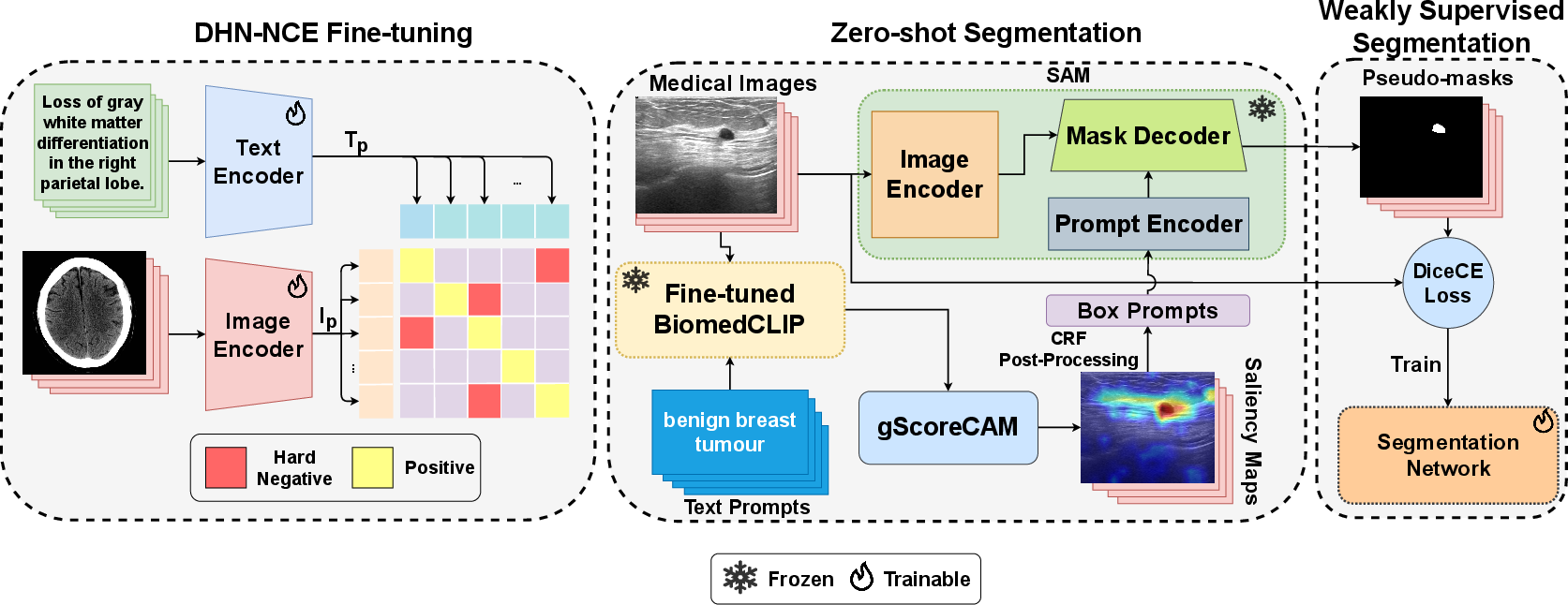
Figure 1: An overview of the proposed MedCLIP-SAM framework.
Efficient BiomedCLIP Fine-Tuning
Decoupled Hard Negative Noise Contrastive Estimation Loss
The DHN-NCE loss combines InfoNCE with hard negative sampling, optimizing medical image-specific tasks by focusing on closely related samples. The elimination of the positive term in the denominator enhances learning efficiency, accommodating smaller batch sizes, essential for sparse medical datasets.
Fine-Tuning Implementation
Using the MedPix dataset, BiomedCLIP is fine-tuned with the DHN-NCE loss, supported by Vision Transformer and PubMedBERT encoders. Images undergo preprocessing, normalization, and a careful train-validation split, ensuring robust learning and retrieval capacities across medical imaging contexts.
Zero-Shot and Weakly Supervised Segmentation
Zero-Shot Segmentation Strategy
Using BiomedCLIP and gScoreCAM, initial segmentation masks are produced, subsequently refined by SAM to establish pseudo-masks. This approach demonstrates versatility across various modalities, including breast ultrasound, brain MRI, and lung X-ray datasets.
Weakly Supervised Refinement
Weakly supervised segmentation exploits pseudo-masks to train Residual UNet models, aiming to enhance segmentation quality further. The effectiveness varies across imaging modalities, improving results notably within X-ray lung segmentation contexts.
Experimental Results
Through comprehensive validation, MedCLIP-SAM exhibits notable accuracy across retrieval and segmentation tasks. Ablation studies confirm gScoreCAM's superiority over GradCAM in generating bounding-box prompts for SAM. Fine-tuned BiomedCLIP models consistently outperform baselines across metrics such as IoU, DSC, and AUC.
Discussion
MedCLIP-SAM emerges as a versatile framework for universal radiological segmentation, leveraging foundation models to facilitate interactive, text-prompt-based anatomical identification. Its innovative DHN-NCE loss contributes substantially to model efficiency and performance. Future directions include exploring MedSAM integration and expanding test scenarios to encompass diverse medical imaging applications.
Conclusion
MedCLIP-SAM integrates state-of-the-art foundation models to offer a novel approach to medical image segmentation, demonstrating strong performance and adaptability across various tasks and modalities. Its contributions pave the way for enhanced clinical applications, fostering interactive AI assistance in medical imaging.


