- The paper introduces the CMTA framework that integrates pathological images and genomic profiles to enhance survival prediction with cross-modal attention.
- It employs dual encoder-decoder architectures and Nystrom self-attention, ensuring efficient extraction and translation of intra- and inter-modal features.
- Experimental results on TCGA datasets show significant improvements over baselines, affirming the framework’s effectiveness for risk stratification.
Cross-Modal Translation and Alignment for Survival Analysis: Technical Summary
Introduction
The paper introduces the Cross-Modal Translation and Alignment (CMTA) framework for survival analysis, targeting the integration of pathological images and genomic profiles. The motivation stems from the limitations of prior multi-modal fusion approaches, which either ignore intrinsic cross-modal correlations or discard modality-specific information. CMTA is designed to extract, translate, and align complementary information across modalities, thereby enhancing the discrimination power for survival prediction tasks.
CMTA Framework Architecture
CMTA employs two parallel encoder-decoder structures, one for pathology and one for genomics. Each encoder utilizes self-attention mechanisms (approximated via Nystrom attention for computational efficiency) to extract intra-modal representations. The decoders translate cross-modal information, ensuring that modality-specific and cross-modal features are both leveraged.
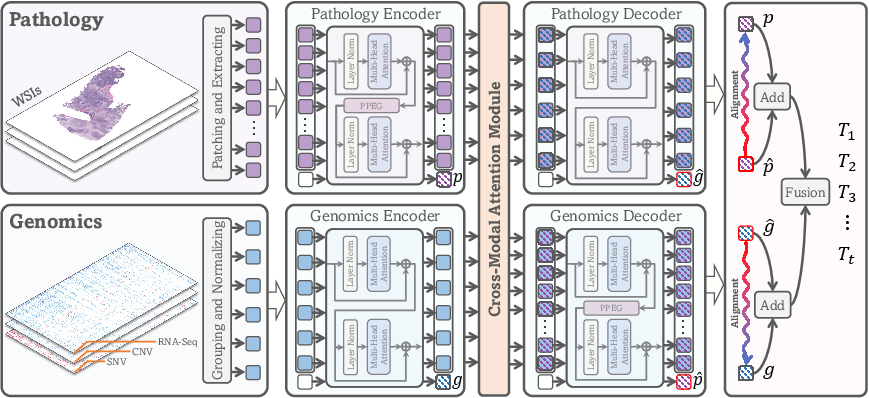
Figure 1: The CMTA framework with parallel encoder-decoder structures for pathology and genomics, cross-modal attention, and feature fusion for survival prediction.
A cross-modal attention module is situated between the encoders and decoders, serving as the information bridge to facilitate interaction and correlation discovery between modalities. This module computes attention maps that quantify the association between pathology and genomics tokens, enabling the extraction of genomics-related information from pathology and vice versa.
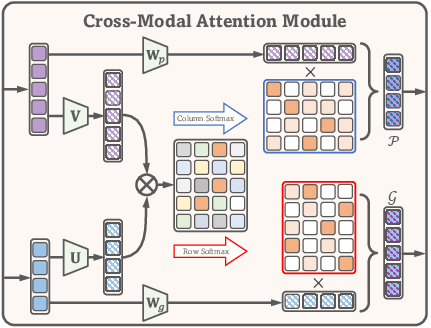
Figure 2: The cross-modal attention module, which computes bidirectional attention maps to capture inter-modality correlations and facilitate information transfer.
Methodological Details
Data Processing and Feature Extraction
- Pathological Images: WSIs are partitioned into 512×512 patches, features are extracted via ResNet-50, and embedded into d-dimensional vectors.
- Genomic Profiles: RNA-seq, CNV, and SNV data are grouped by biological function and embedded similarly.
Encoder Design
- Pathology Encoder: Incorporates a learnable class token and two self-attention layers, with a Pyramid Position Encoding Generator (PPEG) to model spatial correlations.
- Genomics Encoder: Mirrors the pathology encoder but omits the PPEG due to the non-spatial nature of genomic data.
Cross-Modal Attention
The cross-modal attention module computes two attention maps:
- Hp: Genomics-to-pathology associations.
- Hg: Pathology-to-genomics associations.
These maps are used to extract cross-modal features, which are then translated by the respective decoders into cross-modal representations (p^ and g^).
Feature Alignment and Fusion
Cross-modal representations are aligned to intra-modal representations using an L1 norm constraint, with tensor detachment to enforce unidirectional optimization. The final survival prediction is made by fusing intra-modal and cross-modal representations via concatenation and an MLP, with outputs corresponding to discrete time intervals.
Experimental Results
CMTA was evaluated on five TCGA datasets (BLCA, BRCA, GBMLGG, LUAD, UCEC) using 5-fold cross-validation. The primary metric was the concordance index (c-index).
- CMTA consistently outperformed state-of-the-art single-modal and multi-modal baselines.
- Notable improvements over MCAT (previous SOTA) were observed, with c-index increases ranging from 0.89% to 6.39% across datasets.
- Ablation studies demonstrated the necessity of the cross-modal attention module, alignment constraints, and tensor detachment. Removing any of these components resulted in significant performance degradation.
Survival Stratification and Statistical Validation
Patients were stratified into low and high risk groups based on CMTA-predicted risk scores. Kaplan-Meier analysis and logrank tests confirmed statistically significant separation (p<0.05) between groups across all datasets.
Implementation Considerations
- Computational Efficiency: Nystrom attention is critical for handling large patch sets in WSIs.
- Alignment Loss: L1 norm was empirically superior to MSE, KL divergence, and cosine similarity for most datasets.
- Optimization: Tensor detachment during alignment loss computation is essential to prevent collapse to redundant shared information.
- Batching: Due to gigapixel image sizes, mini-batch optimization is infeasible; discrete time interval modeling is used instead.
Implications and Future Directions
CMTA demonstrates that explicit modeling of cross-modal correlations and translation of complementary information yields superior survival prediction performance. The framework is extensible to other multi-modal biomedical tasks, such as radiology-pathology-genomics integration or multi-omics fusion. Future work may explore:
- Scaling to additional modalities (e.g., clinical text, proteomics).
- More sophisticated alignment objectives (e.g., adversarial or contrastive losses).
- Efficient training strategies for ultra-large datasets.
Conclusion
CMTA provides a robust and principled approach for multi-modal survival analysis, leveraging cross-modal translation and alignment to fully exploit complementary information. The framework sets a new benchmark for survival prediction on TCGA datasets and offers a template for future multi-modal learning systems in computational pathology and genomics.



