- The paper presents an empirical evaluation showing that mix-based augmentation techniques consistently outperform traditional methods in physiological time series classification.
- It details the use of mixup, cutmix, and manifold mixup across six datasets, demonstrating improved accuracy and effective handling of class imbalances.
- Results indicate that these methods enhance feature representation and model interpretability via vicinal risk minimization and t-SNE visualizations.
Empirical Evaluation of Mix-based Data Augmentation Methods in Physiological Time Series Data
Introduction
Data augmentation is an established technique to improve model generalization by simulating variety within datasets. Traditionally used approaches in time series data involve transformations such as jittering, scaling, and permutation, which aim to preserve label invariance. However, many of these methods are dataset-dependent and may introduce alterations detrimental to physiological signal integrity. This paper explores mix-based augmentation techniques—mixup, cutmix, and manifold mixup—originating from the computer vision domain, assessing their utility within physiological time series classification tasks.

Figure 1: The overview of mix-based augmentation procedure for physiological time series classification.
Methodology
The study introduces three mix-based augmentations:
- Mixup: Generates virtual samples by linear interpolations of inputs and labels.
- Cutmix: Replaces segments of data from one input with segments from another.
- Manifold Mixup: Applies mixing at the feature map level within neural networks.
These methods are applied across six diverse physiological datasets, with results compared against baseline models and traditional augmentation techniques. Mix-based methods are of particular interest due to their independence from dataset-specific knowledge and parameter tuning.

Figure 2: Illustrations of traditional time series data augmentations.
Empirical Evaluation
Performance Across Datasets: In experiments, mix-based augmentation methods consistently outperformed traditional augmentation techniques. Mixup and its variants showed improved accuracy and less dependency on expert parameter tuning. Specifically, cutmix and manifold mixup yielded superior results across multiple datasets such as PTB-XL, Apnea-ECG, Sleep-EDF, MMIDB, PAMAP2, and UCI-HAR.
Case Analysis with PTB-XL Dataset: The PTB-XL dataset, characterized by class imbalance, was used to further profile these methods. Mix-based augmentations not only improved classification metrics but also enhanced predictions for minority classes when paired with a class-balanced sampler (Figure 3).
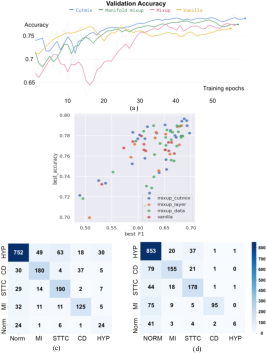
Figure 3: (a): The PTB-XL validation accuracy computed after each training epoch, with baseline and variations of mixup. (b): The scatter plot of validation accuracy against F1 score for all 80 profiling experiments on PTB-XL dataset.
Feature Representation and Interpretation
t-SNE visualizations illustrated that mix-based methods facilitate better separability in feature space, thereby enhancing model interpretability and generalization (Figure 4). This supports the notion that vicinal risk minimization inherent in mixup augments class-distinct feature representations.

Figure 4: The t-SNE visualizations of cutmix and vanilla settings after training on PTB-XL.
Conclusion
This study underscores the efficacy of mix-based data augmentation techniques in time series classification. By leveraging methods that are agnostic to specific dataset attributes, these techniques offer robust performance improvements without extensive parameterization. Future research will focus on integrating these mix-based methods with traditional augmentations and extending their applicability to frequency-domain features in time series data.





