Large Language Models for Healthcare Data Augmentation: An Example on Patient-Trial Matching (2303.16756v2)
Abstract: The process of matching patients with suitable clinical trials is essential for advancing medical research and providing optimal care. However, current approaches face challenges such as data standardization, ethical considerations, and a lack of interoperability between Electronic Health Records (EHRs) and clinical trial criteria. In this paper, we explore the potential of LLMs to address these challenges by leveraging their advanced natural language generation capabilities to improve compatibility between EHRs and clinical trial descriptions. We propose an innovative privacy-aware data augmentation approach for LLM-based patient-trial matching (LLM-PTM), which balances the benefits of LLMs while ensuring the security and confidentiality of sensitive patient data. Our experiments demonstrate a 7.32% average improvement in performance using the proposed LLM-PTM method, and the generalizability to new data is improved by 12.12%. Additionally, we present case studies to further illustrate the effectiveness of our approach and provide a deeper understanding of its underlying principles.
Sponsor
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
What is this paper about?
This paper shows how LLMs—the kind of AI that understands and writes text—can help match patients to the right clinical trials. The authors focus on keeping patient information private while making the “rules” of a trial easier for computers to understand. Their approach, called LLM-PTM, rewrites trial rules in many clear ways so a matching system can do a better job, without sending sensitive patient details to the AI.
What questions were the researchers trying to answer?
They set out to explore a few simple questions:
- Can an AI that’s good with language make it easier to match patient records to a trial’s rules?
- Can this be done while protecting patient privacy?
- Will these AI-made versions of trial rules help the system work better on new, unseen trials?
How did they do it? (Simple explanation of the methods)
Think of patient-trial matching like a dating app for healthcare: a patient’s medical history needs to “match” the list of who can join a trial (inclusion criteria) and who cannot (exclusion criteria).
The problem: patient records (EHRs) and trial rules are written differently, using different words and formats. That makes matching hard.
What they built:
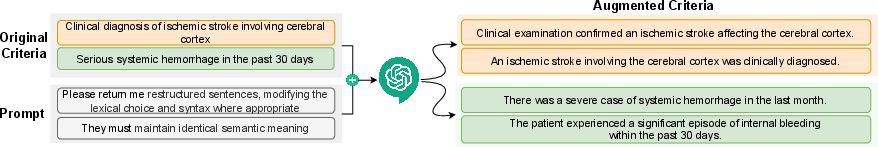
- Step 1: Rewriting the rules (data augmentation). The LLM takes each trial rule and rewrites it into several clear, slightly different versions that mean the same thing. This is like making multiple practice questions with the same answer so a student (the matching model) learns better.
- Privacy move: They don’t send raw patient details to the AI. Instead, they use “desensitized” (de-identified) hints so the AI knows how to rewrite the rules in helpful, privacy-safe ways.
- They guide the AI with chain-of-thought prompts (instructions that help the AI reason step-by-step) to keep the meaning identical while making the wording machine-friendly.
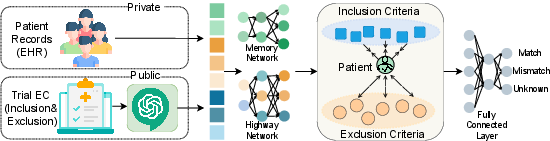
- Step 2: Turning text into numbers (embeddings). Computers understand numbers better than words, so they use a model called Clinical BERT (a program trained on lots of hospital notes) to convert both:
- Patient records into numerical “summaries” that remember the order of visits (think: a timeline memory).
- Trial rules into numerical “summaries” too.
- Step 3: Teaching the matcher. They train a model to:
- Predict whether a patient matches each rule: match, mismatch, or unknown.
- Treat inclusion and exclusion rules differently (like magnets: pull together for inclusion, push apart for exclusion), so the model pays attention to words like “must” vs. “must not.”
- Step 4: Testing. They used data from six stroke trials and 825 patients. They created many patient–rule pairs and checked how well the system matched them at two levels: rule-by-rule and whole-trial.
What did they find, and why does it matter?
Main results:
- Better accuracy: On average, their LLM-PTM approach improved performance by 7.32% compared to not using the AI-written rule versions.
- Stronger on new data: When tested on trials the model hadn’t seen before, performance improved by 12.12%. That means the system is more flexible and can handle new trials better.
- More consistent across trials: Some trials were harder than others, but the LLM-based rewrites helped smooth out those difficulties.
- Real examples helped explain why: The AI rewrites turned short, tricky phrases (like “acute ischemic stroke”) into clearer versions (“a sudden blockage of blood flow to the brain”), which made it easier for the model to match the right patients.
Why it matters:
- Clinical trials often struggle to find the right participants quickly. Clearer, AI-augmented rules help computers find matches faster and more accurately, which can speed up medical research and help patients access treatments.
Why is this important for the future?
- Faster, fairer recruitment: Better matching can reduce delays in trials, potentially bringing new treatments to patients sooner.
- Privacy-aware AI in healthcare: This shows a practical way to use powerful AIs without exposing private patient data.
- Reusable idea: The same “rewrite-and-learn” approach could help with other healthcare tasks where important text is written in different ways (like insurance rules, referral guidelines, or hospital policies).
- Path to safer, smarter systems: By making text clearer for machines while keeping meaning the same, AI can assist doctors and researchers without replacing them—supporting better decisions and outcomes.
Practical Applications
Practical Applications of “LLMs for Healthcare Data Augmentation: An Example on Patient-Trial Matching”
Below are actionable applications that flow directly from the paper’s findings (notably a 7.32% average performance gain and 12.12% generalizability improvement) and its innovations (privacy-aware LLM-based augmentation of trial criteria, inclusion/exclusion-aware contrastive training, and ClinicalBERT-based embeddings with memory networks). Each item indicates relevant sectors, potential tools/workflows, and key assumptions/dependencies that could affect feasibility.
Immediate Applications
- Patient–trial matching uplift via LLM-driven criteria augmentation
- Sectors: healthcare providers, CROs/pharma sponsors, EHR vendors, clinical research IT
- What: Use the LLM-PTM augmentation to generate machine-friendly paraphrases of inclusion/exclusion criteria (ECs), retrain existing PTM models to realize the reported performance and generalizability gains on current trials.
- Tools/workflows: “Criteria Augmentation” microservice; ClinicalBERT/COMPOSE retraining pipeline; prompt templates with chain-of-thought and desensitized patient features; MLOps for model versioning.
- Assumptions/dependencies: High-quality redaction/desensitization; access to LLMs (API or on-prem); site IRB approval; availability of structured EHR (ICD/SNOMED, RxNorm, CPT) and ground-truth labels.
- EHR-integrated pre-screening assistant (human-in-the-loop)
- Sectors: hospital research offices, principal investigators, EHR vendors
- What: Rank prospective patients for ongoing trials using augmented criteria embeddings; coordinators review and confirm.
- Tools/workflows: Pre-screening dashboard; eligibility score with inclusion vs exclusion highlights; queue triage; audit logs.
- Assumptions/dependencies: HIPAA compliance, data use agreements, workflow integration (e.g., FHIR ResearchStudy/Patient); clinical validation at the site.
- Trial feasibility and site selection analytics
- Sectors: CROs, pharma sponsors, health system administrators
- What: Use improved matching to estimate eligible patient volumes across sites and time for feasibility assessments and site selection.
- Tools/workflows: Cohort estimation pipeline; accrual projections; dashboards.
- Assumptions/dependencies: Representative EHR coverage; historical recruitment data; careful handling of domain shift (paper focused on stroke).
- Authoring and quality control assistant for eligibility criteria
- Sectors: medical writing, regulatory affairs, protocol design teams
- What: Rephrase ECs into unambiguous, machine- and human-friendly language; surface negations and contradictions to reduce misinterpretation.
- Tools/workflows: EC rewrite/checker tool; inclusion/exclusion contrast-focused linting; semantic equivalence checks.
- Assumptions/dependencies: Sponsor and IRB acceptance; traceability of edits; regulatory documentation needs.
- Terminology harmonization layer between EHR and trial criteria
- Sectors: software/AI, standards (CDISC, HL7), EHR vendors
- What: Generate synonym/normalization maps (e.g., ischemic stroke variants) to bridge protocol text and coded EHR concepts.
- Tools/workflows: Mapping service to ICD-10/SNOMED CT, RxNorm, CPT/LOINC; curation UI.
- Assumptions/dependencies: Vocabulary licensing; human-in-the-loop curation for correctness.
- Privacy-aware LLM augmentation pattern for clinical NLP
- Sectors: healthcare IT security, compliance, clinical NLP teams
- What: Operationalize desensitization/redaction and zero-trust interactions with LLMs for augmentation tasks (beyond PTM).
- Tools/workflows: DLP/redaction service; on-prem or VPC-hosted LLMs; privacy audit trails and prompts catalogs.
- Assumptions/dependencies: BAAs with vendors; measurable re-identification risk controls; prompt leakage prevention.
- Clinical NLP data efficiency: semi-automated labeling and augmentation
- Sectors: academia, software/AI
- What: Reduce manual labeling by generating diverse, semantically faithful EC paraphrases and “unknown” class examples to balance datasets.
- Tools/workflows: Augmentation pipelines; active learning loops; evaluation harnesses.
- Assumptions/dependencies: Label noise monitoring; task-specific prompt tuning; robust test sets.
- Recruiter/call-center screening support with patient-friendly EC explanations
- Sectors: CROs, patient advocacy groups
- What: Translate technical ECs into lay language to speed preliminary screenings and reduce miscommunication.
- Tools/workflows: Explanation generator; script suggestions for coordinators; consistency checks.
- Assumptions/dependencies: Medical-legal review; consistent terminology; multilingual support as needed.
- Academic benchmarking and reproducibility kits
- Sectors: academia, open-source community
- What: Release augmentation recipes, code, and datasets (where permissible) for PTM benchmarking with LLM-PTM vs baselines.
- Tools/workflows: Reproducible training pipelines; dataset cards; seeds/metrics parity.
- Assumptions/dependencies: Data licensing/IRB constraints; generalizability beyond stroke.
Long-Term Applications
- Regulated, end-to-end automated trial matching as clinical decision support
- Sectors: healthcare providers, EHR vendors, regulators (FDA/EMA)
- What: Near real-time eligibility scoring with explanations embedded in clinician workflows; human oversight and auditability.
- Tools/workflows: SaMD-grade pipeline; FHIR-based integration (ResearchStudy/PlanDefinition/Task); monitoring for drift and bias.
- Assumptions/dependencies: Prospective clinical validation; regulatory clearance; reliability and fairness audits.
- Federated, privacy-preserving multi-site PTM training
- Sectors: health-system consortia, academic medical centers
- What: Train PTM models across institutions using local LLM augmentation and federated learning to avoid data centralization.
- Tools/workflows: Secure aggregation; DP/HE options; site-specific augmenters.
- Assumptions/dependencies: Governance agreements; compute at edge; harmonized coding standards.
- Adaptive eligibility optimization to improve accrual and diversity
- Sectors: pharma sponsors, regulators, bioethics committees
- What: Simulate and propose data-driven EC relaxations or clarifications to boost inclusion of underrepresented groups without compromising safety.
- Tools/workflows: Scenario EC generator; safety constraints; subgroup eligibility analytics.
- Assumptions/dependencies: Clinical risk modeling; policy guidance; oversight for ethical implications.
- Cross-disease and multilingual expansion
- Sectors: life sciences (oncology, cardiology, rare diseases), global trial operations
- What: Generalize LLM-PTM across specialties and languages to support international trials.
- Tools/workflows: Domain-adapted encoders (e.g., OncologyBERT); multilingual LLMs; expanded ontologies.
- Assumptions/dependencies: Domain-specific labeled data; localization; varied privacy regimes.
- Patient-facing trial discovery with personalized, explainable eligibility
- Sectors: digital health, patient engagement
- What: Consumer apps that summarize likely eligibility, surface missing information, and guide pre-consent education.
- Tools/workflows: Lay-language explanation engines; risk/benefit summaries; referral workflows to sites.
- Assumptions/dependencies: Safety/liability guardrails; accessibility/readability; misinformation safeguards.
- Standards evolution toward computable trial criteria
- Sectors: standards bodies (CDISC, HL7), regulatory science
- What: Translate LLM-normalized ECs into CDISC/HL7 FHIR PlanDefinition/Library assets to enable interoperable, computable protocols.
- Tools/workflows: Authoring plugins; mapping pipelines; conformance validators.
- Assumptions/dependencies: Community consensus; backward compatibility; governance.
- Payer utilization management and prior authorization matching
- Sectors: payers/finance, providers
- What: Map clinical indications to coverage criteria using LLM-normalized rules to reduce administrative burden and denials.
- Tools/workflows: Criteria matching engine; explainable decisions; appeals support.
- Assumptions/dependencies: Integration with payer policies; fairness and non-discrimination audits.
- Trial operations forecasting and resource optimization
- Sectors: clinical operations, finance
- What: Use improved matching and generalizability to forecast accrual, guide staffing, and optimize budgets across sites.
- Tools/workflows: Predictive dashboards; CTMS integration; scenario planning.
- Assumptions/dependencies: Reliable historical data; model calibration; operational buy-in.
- Pharmacovigilance and post-market study enrollment
- Sectors: safety surveillance, registries
- What: Identify eligible patients for registries/REMS using precise inclusion/exclusion semantics and EHR signals.
- Tools/workflows: EHR alerting; outreach automation; consent tracking.
- Assumptions/dependencies: Real-time data feeds; consent management; alert fatigue controls.
- Bias and fairness auditing of eligibility language
- Sectors: ethics/DEI, regulatory science
- What: Detect exclusionary phrasing and simulate subgroup impacts; recommend alternative EC wording.
- Tools/workflows: Audit reports; counterfactual paraphrase generator; fairness metrics.
- Assumptions/dependencies: Access to demographic attributes; governance for policy actions.
- On-prem privacy-certified LLMs for clinical augmentation
- Sectors: IT vendors, large health systems
- What: Deploy fine-tuned, domain-specific LLMs with privacy certifications to avoid external data transfer.
- Tools/workflows: Model serving stack; RLHF for clinical style; red team testing; monitoring.
- Assumptions/dependencies: GPU/TPU capacity; model licensing; TCO considerations.
- Global, cross-lingual recruitment and outreach
- Sectors: public health, NGOs, global CROs
- What: Normalize and match ECs across languages/health systems to widen access to trials.
- Tools/workflows: Translation + normalization pipelines; locale-specific vocabularies; outreach tooling.
- Assumptions/dependencies: Language coverage; compliance with local regulations; cultural adaptation.
Collections
Sign up for free to add this paper to one or more collections.