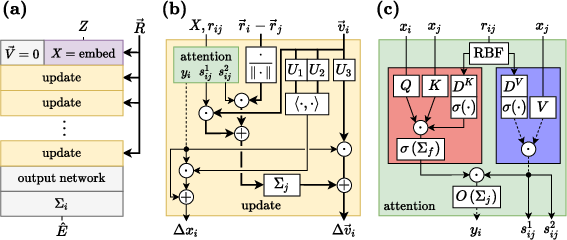
- The paper introduces TorchMD-NET, a novel equivariant Transformer that outperforms conventional methods on datasets like MD17, ANI-1, and QM9.
- The modified attention mechanism incorporates edge attributes and atomistic embeddings, enhancing the capture of spatial relationships in molecules.
- Ablation studies highlight the critical role of embedding and update layers in boosting accuracy while maintaining competitive computational efficiency.
Introduction
The accurate prediction of quantum mechanical properties has traditionally been constrained by a trade-off between accuracy and computational efficiency. TorchMD-NET introduces a novel equivariant Transformer (ET) architecture that seeks to balance these factors, outperforming state-of-the-art methods on significant datasets such as MD17, ANI-1, and QM9 in terms of both precision and computational demand. This architecture is particularly centered on the attention mechanism inherent in Transformer models, enabling it to surpass previous approaches through a refined representation of atomic features.
Methods
TorchMD-NET employs a modified self-attention mechanism tailored for the molecular domain, where data is naturally structured as a graph. This involves three primary architectural components: an embedding layer, a modified attention mechanism, and an update layer.
Experiments and Results
TorchMD-NET's performance was evaluated on several prominent datasets, demonstrating its efficacy across different molecular configurations:
- QM9: The ET model excelled in predicting various quantum-chemical properties with superior mean absolute errors (MAE) when compared to prior models like SchNet, PhysNet, and DimeNet++.
- MD17: Excelling particularly in predicting molecular dynamics with accurate force and energy predictions, the model's performance is remarkable considering the limited data regime it was trained on.
- ANI-1: TorchMD-NET exhibited impressive accuracy on large datasets with off-equilibrium conformations, underlying its strength in learning from diverse molecular geometries.
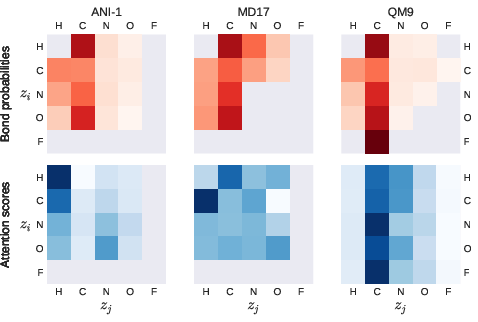
Figure 2: Depiction of bond probabilities and attention scores across different datasets.
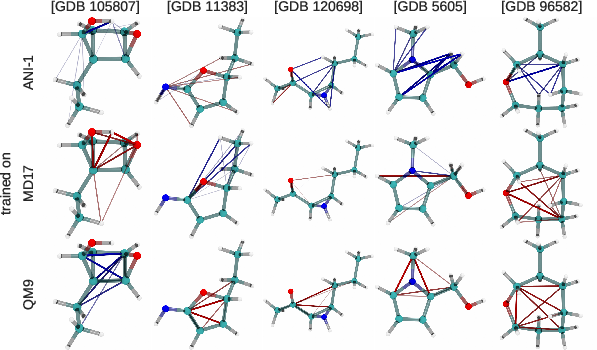
Figure 3: Visualization of attention scores on molecules from the QM9 dataset, reflecting positive and negative attention dynamics.
Attention Weight Analysis
A critical innovation of TorchMD-NET is its attention weight analysis, providing insights into the underlying mechanisms of the model. The attention mechanism unveils how the model recognizes and accentuates various atom-atom interactions, distinctively influenced by the dataset characteristics. Hydrogen atoms, for example, receive differentiated attention based on the nature of the dataset, highlighting configurational dependency in the model's focus.
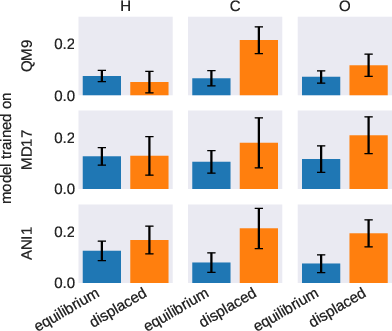
Figure 4: Averaged attention weights indicating sensitivity to atomic displacements.
Ablation and Computational Efficiency
Ablation studies underscore the importance of each component, with neighbor embedding layers and equivariant features contributing significantly to the model’s accuracy. Despite its enhanced size due to additional parameters, TorchMD-NET maintains competitive computational efficiency, rivaling smaller architectures in inference speed while offering superior accuracy.
Discussion
TorchMD-NET represents a significant progression in the domain of quantum mechanical property prediction, leveraging an equivariant Transformer architecture that integrates learned atomistic features with attention-based dynamics. The architecture delivers state-of-the-art performance on molecular datasets, asserting the importance of configurational diversity and the role of attention mechanisms in capturing complex atomic interactions.
Conclusion
TorchMD-NET has spearheaded advancements in neural network-based molecular potentials, offering an equivariant approach that harmonizes accuracy with computational efficiency. Its capacity to attend to specific atomic interactions opens new avenues for refining molecular simulations, with implications for improved predictions in materials science, chemistry, and drug discovery. Future developments may extend this approach to even broader classes of quantum properties, enhancing its applicability across diverse scientific domains.