- The paper introduces a novel CNN architecture (FSConv) for patch-level Gleason grading, achieving a quadratic Cohen’s kappa of 0.77.
- It fine-tunes the FSConv model for cribriform pattern detection in Gleason grade 4 patches, yielding an ROC AUC of 0.82.
- The system integrates patch-level outputs into whole slide assessments using an MLP, achieving a biopsy scoring kappa of 0.81.
Automatic End-to-End System for Prostate Histology Grading
Introduction
The paper "Going Deeper through the Gleason Scoring Scale: An Automatic end-to-end System for Histology Prostate Grading and Cribriform Pattern Detection" presents an innovative approach to enhance the diagnosis and analysis of prostate cancer—specifically through histology biopsies—with an automatic system. Prostate cancer remains one of the most prevalent malignancies among men worldwide. Accurate grading and scoring are critical for prognosis and subsequent treatment decisions, predominantly relying on Gleason grading. However, the manual analysis of histology samples is labor-intensive, subjective, and prone to variability, imposing significant workloads on pathologists.
Technological advancements in digitization and computer vision now allow for automating some aspects of histology analysis. This paper aims to develop and validate a comprehensive end-to-end system leveraging deep learning techniques to bolster pathologists' analyses, focusing not just on Gleason grading but also on detecting specific patterns within Gleason grade 4—such as the prognostically significant cribriform pattern.
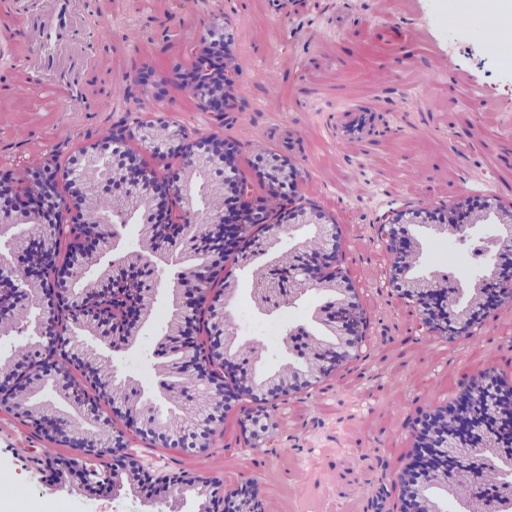
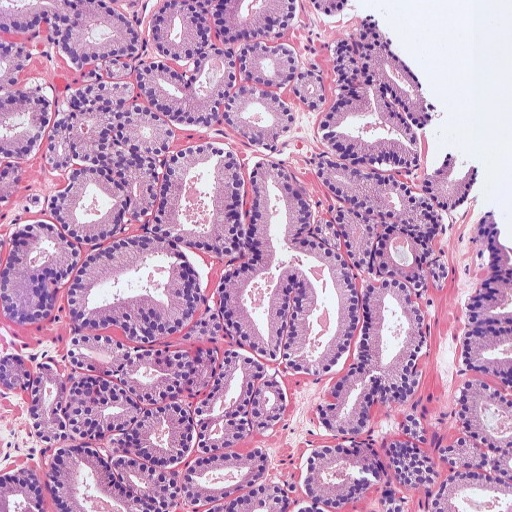
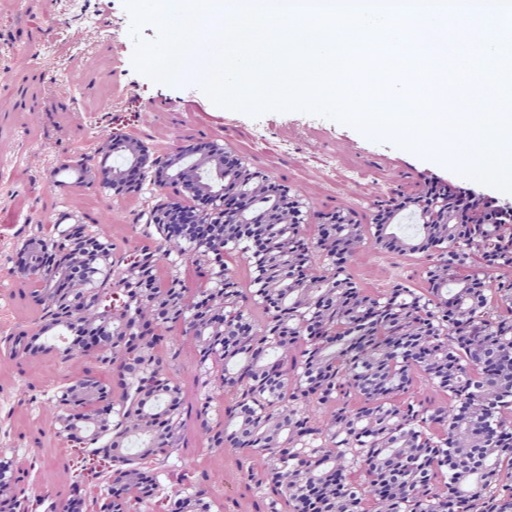
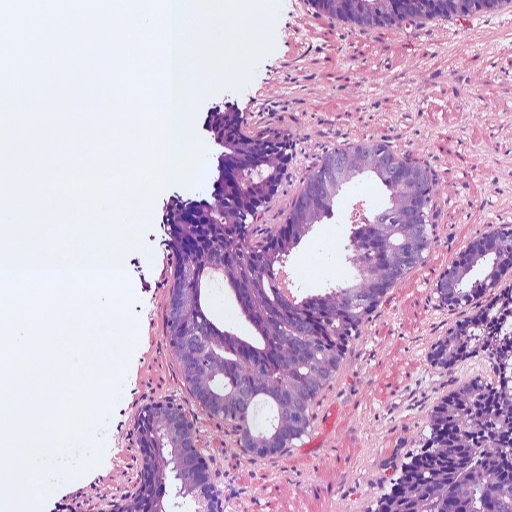
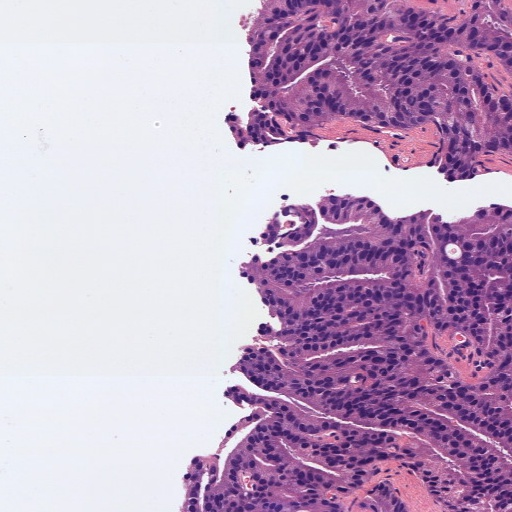
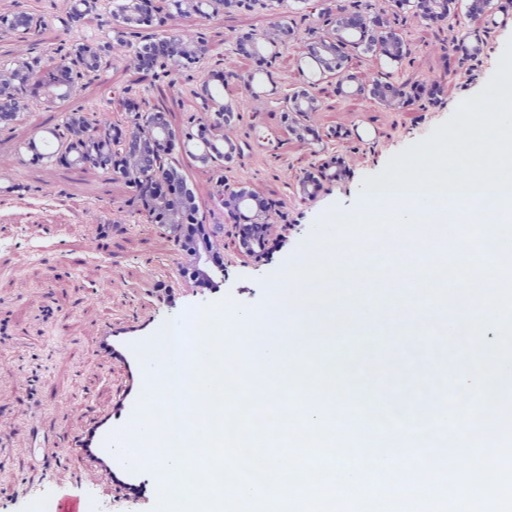
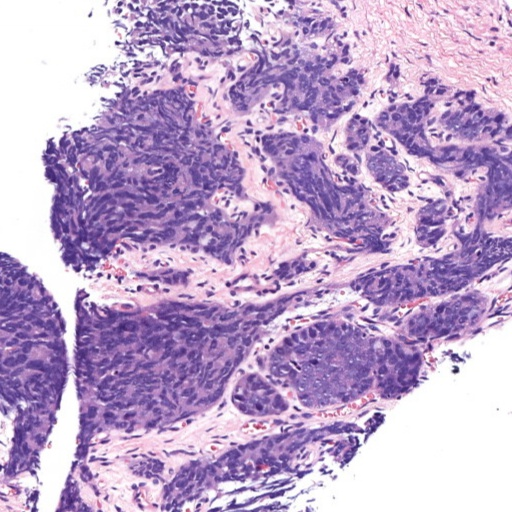
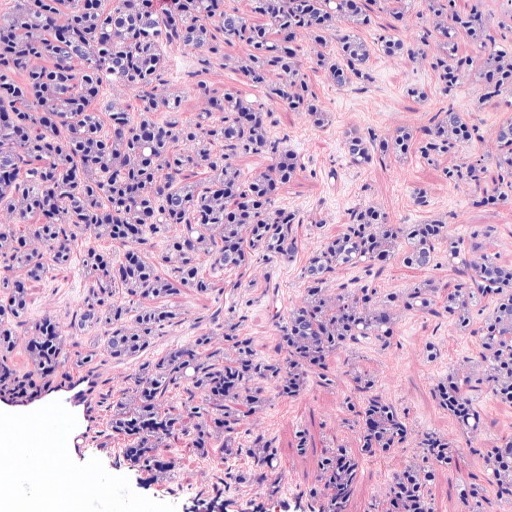
Figure 1: Patches of H&E histology samples presenting different Gleason patterns.
Methods
Patch-Level Gleason Grading
The core of the proposed system is a convolutional neural network (CNN) designed to predict Gleason grades at the patch level. The authors introduce a self-designed CNN architecture named FSConv, characterized by three simple convolutional layers and employing max-pooling for dimensional reduction (Figure 2). The system adopts global-max pooling as a top model to mitigate issues related to the variability of tissue pattern location and size within histology patches, thus reducing the model's complexity and minimizing overfitting risks.
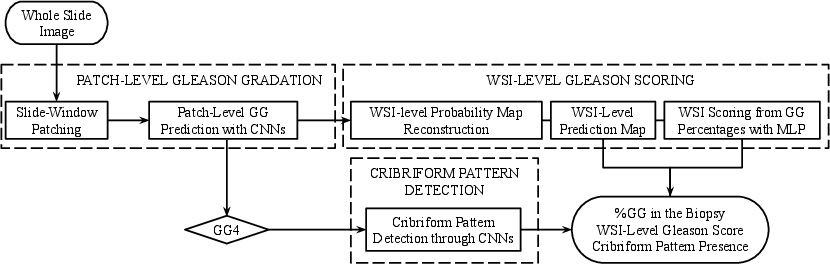
Figure 2: Flowchart in which the different blocks of our system are presented.
The detection of cribriform patterns in Gleason grade 4 patches was addressed by fine-tuning the FSConv model initially trained for Gleason grade prediction. Focusing on the complexity of this task and the limited availability of cribriform samples, the fine-tuning approach allowed for optimizing the system without requiring extensive computational resources or adjusting a vast array of parameters.
Whole Slide Image Gleason Scoring
The system reconstructs biopsy-level prediction maps based on patch-level outputs through bi-linear interpolation, which serves to compute the percentage of each Gleason grade within the tissue. A multi-layer perceptron (MLP) is developed to predict the biopsy-level Gleason score, factoring in both the proportion and the severity of the grades present.
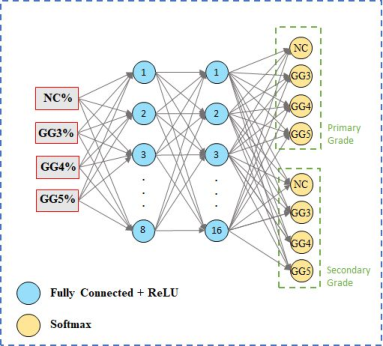
Figure 3: Proposed Multi-Layer Perceptron (MLP) for the whole slide image Gleason scoring.
Results
The system was rigorously validated on the SICAPv2 database, composed of 182 whole slide images. The FSConv network achieved a strong agreement with pathologist annotations, with a quadratic Cohen’s kappa of 0.77 for patch-level grading, surpassing previous benchmarks. Additionally, the cribriform pattern detection task yielded an ROC curve area of 0.82, affirming the reliability of the model in medical contexts.
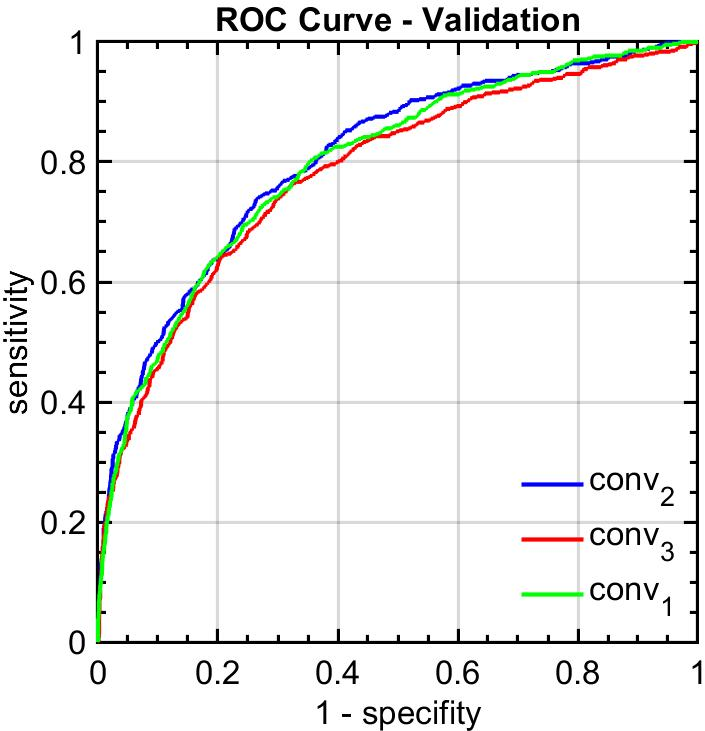
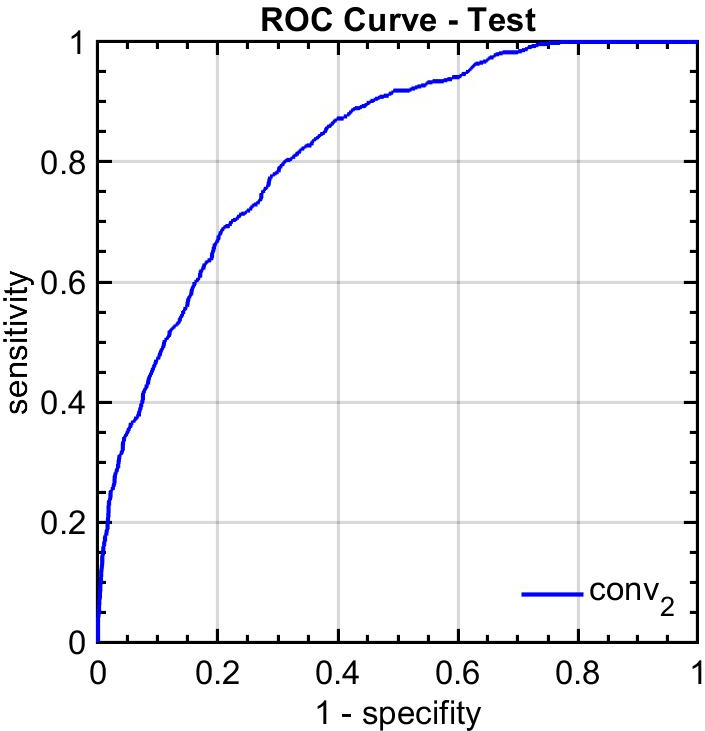
Figure 4: ROC curves obtained for cribriform pattern detection in samples with Gleason grade 4.
Moreover, the biopsy Gleason scoring achieved a quadratic Cohen’s kappa of 0.81, showcasing the MLP model’s fidelity in simulating pathologists' decision-making processes more accurately than previous simpler models.
Discussion
This system represents a significant advancement in automating prostate biopsy grading, enabling more efficient and accurate assessments which could reduce pathologist workload and variability. Importantly, this is the first paper addressing the automatic detection of cribriform patterns—a critical factor in assessing severe prostate cancer cases. Future work aimed at refining the CNN architecture could further integrate low and high-level features for robust pavilion-style training, potentially extending the system’s capability to predict other individual cancerous patterns.
Conclusion
The work detailed in this paper reflects a substantial step forward in prostate histology analysis. The presented end-to-end system exhibits strong potential for practical deployment, promising to support and streamline pathologist workflows. With continual development and enlargement of databases like SICAPv2, combined with increasingly sophisticated deep learning models, the field is poised to significantly enhance diagnostic accuracy and efficiency in prostate and potentially other cancers.













