- The paper introduces a novel EBM-NLP corpus that systematically annotates 5,000 RCT abstracts for PICO elements to support advanced NLP in medicine.
- It details a two-phase annotation strategy combining crowdsourced input and expert validation to ensure high-quality, multi-level annotations.
- The corpus enhances model training for information extraction and evidence synthesis, facilitating better decision-making in clinical practice.
Summary of "A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature" (1806.04185)
Introduction
The paper introduces a comprehensive corpus designed to enhance the application of NLP in evidence-based medicine (EBM). It addresses the need to efficiently process and synthesize the growing body of biomedical literature, specifically focusing on clinical randomized controlled trials (RCTs). By presenting EBM-NLP, a new annotated dataset, the researchers aim to bridge the gap between the complexities of medical text and the capabilities of current NLP systems. The dataset is tailored to facilitate the extraction of patient populations, interventions, and outcomes, commonly referred to as the PICO elements, thereby supporting the aims of EBM to use all available evidence in informing patient care.
Two primary research areas are examined: NLP applications in EBM and the role of crowdsourcing in generating NLP resources. Historically, the lack of large, annotated datasets has hampered progress in the NLP domain regarding EBM. Previous efforts relied on small datasets or noisily annotated large corpora. Moreover, while crowdsourcing has shown potential in the domain of simple NLP tasks, its application in the biomedical field, which demands specialized knowledge, has been less explored.
Corpus Development
The EBM-NLP corpus consists of 5,000 annotated abstracts from medical literature, specifically RCTs, focusing on cardiovascular diseases, cancer, and autism. The corpus features multi-level annotations of PICO elements, enabling granular information extraction. Data collection involved recruiting a diverse pool of annotators, ranging from laypersons via Amazon Mechanical Turk to medical experts via platforms like Upwork. The objective was to create a high-quality dataset while managing the trade-off between annotation cost and expertise.
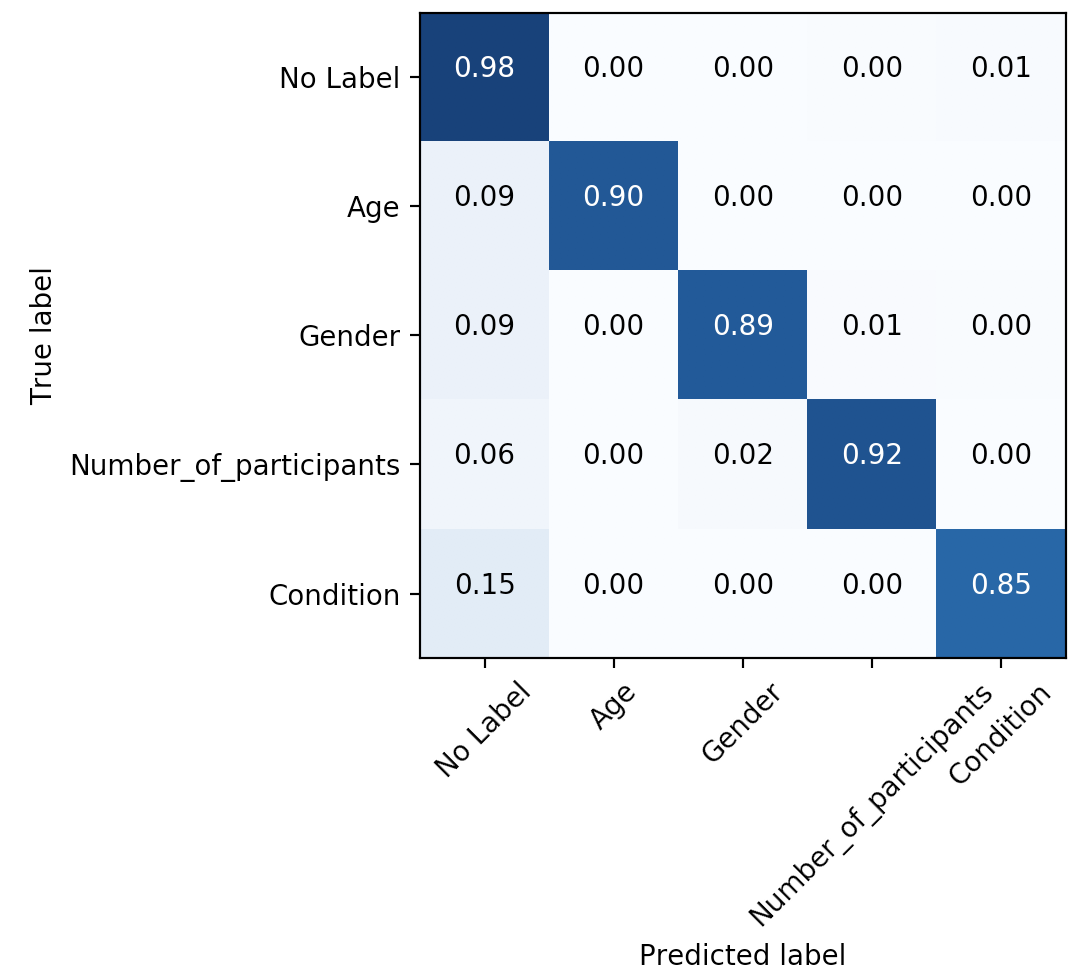
Figure 1: Confusion matrix for token-level labels provided by experts.
Annotation Strategy
The annotation process was structured in two phases. In the first phase, annotators identified text spans relevant to each PIO element. This was followed by a second phase where these spans were annotated in finer detail, including hierarchical labeling aligned with MeSH terminology. The study employed various models to aggregate worker annotations and produce high-confidence labels, such as the HMMCrowd model which leverages sequence data structure.
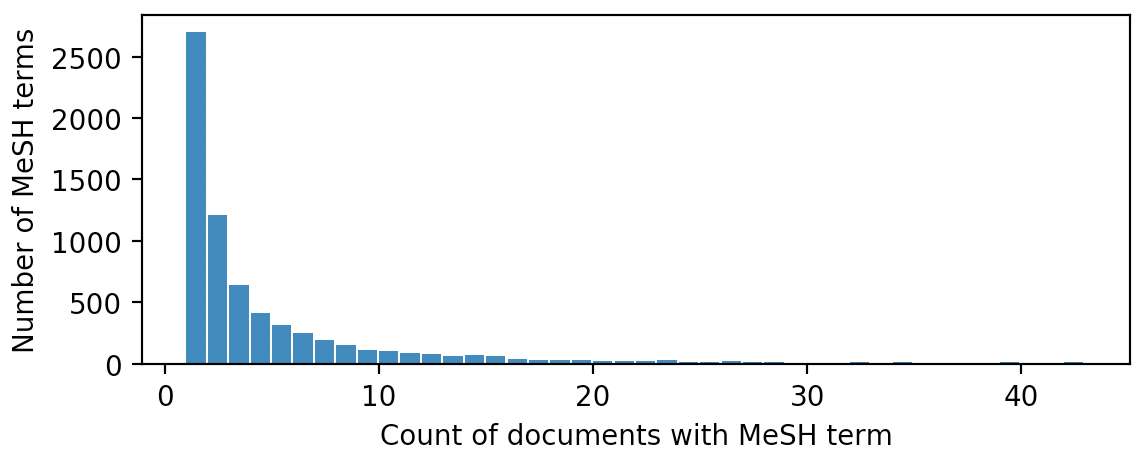
Figure 2: Histogram of the number of documents containing each MeSH term.
Quality and Evaluation
To ensure the reliability of the dataset, the study measured inter-annotator agreement and performed various accuracy assessments. While individual crowdsourced annotations showed variability, aggregated annotations were of comparable quality to expert annotations. The introduction of multiple aggregation strategies improved the final dataset quality, allowing for resilient NLP model training on this complex data.

Figure 3: Span level recall of workers vs. how many documents they annotated (on P).
Implications
The release of this corpus is expected to significantly enhance NLP research in the biomedical domain. It provides a valuable resource for developing and testing models in information extraction, document retrieval, and evidence synthesis. The dataset can enable more precise question-answering systems in EBM, supporting automated approaches to literature review and synthesis.
Conclusion
The EBM-NLP corpus represents a crucial contribution to the field of biomedical NLP by providing a resource primed for facilitating advancements in automatic evidence synthesis and retrieval. Its comprehensive annotations and diverse sourcing promise to support the development of more robust NLP systems in medicine, thereby improving the practice of EBM. Future work may involve expanding the corpus and refining NLP techniques to leverage this resource more effectively. The dataset and baseline methodologies are made publicly available to catalyze further research in this domain.

Figure 4: How many times is each label used per document? (for P).





