- The paper presents CNNCS, a framework that encodes sparse cell locations into compressed vectors and recovers them via L1-norm minimization.
- It integrates deep CNN regression using architectures like AlexNet and ResNet with multi-task learning to enhance detection accuracy.
- The method is theoretically justified using the restricted isometry property and empirically outperforms traditional techniques across multiple datasets.
Deep Convolutional Neural Network and Compressed Sensing for Cell Detection in Microscopy Images
Introduction
The paper presents a novel framework, CNNCS, for cell detection and localization in microscopy images, integrating deep convolutional neural networks (CNNs) with compressed sensing (CS)-based output encoding. The approach addresses the inherent sparsity of cell locations in high-resolution images and the limitations of direct pixel-space classification, particularly the class imbalance and susceptibility to false positives. By encoding cell locations into a compressed vector via random projections and regressing this vector from image data using a CNN, the method leverages L1-norm optimization for robust recovery of cell centroids. This paradigm shift from direct coordinate prediction to compressed signal regression is theoretically justified by the restricted isometry property (RIP) of CS and is empirically validated on multiple benchmark datasets.
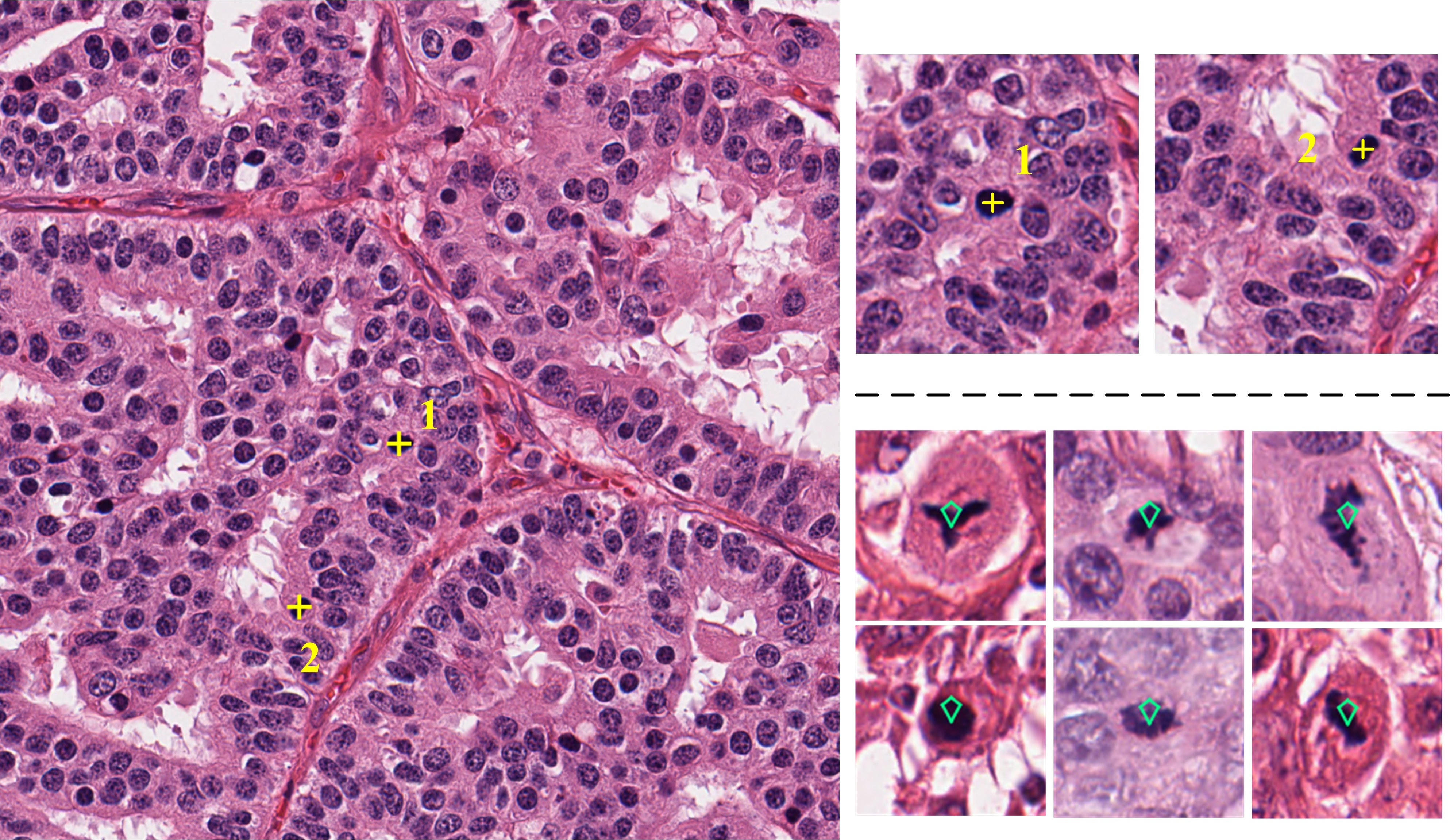
Figure 1: Microscopy image with annotated mitotic cells and examples of mitotic figures.
Methodology
System Architecture
The CNNCS framework comprises three principal stages: (1) cell location encoding via random projection, (2) CNN-based regression to predict the compressed signal, and (3) decoding through L1-norm minimization to recover cell locations. The encoding transforms the sparse binary annotation map of cell centroids into a fixed-length compressed vector, which serves as the regression target for the CNN. Two encoding schemes are proposed: Scheme-1 (reshaping the annotation map) and Scheme-2 (encoding signed distances to multiple observation axes), with Scheme-2 offering significant computational advantages and redundancy for improved detection reliability.
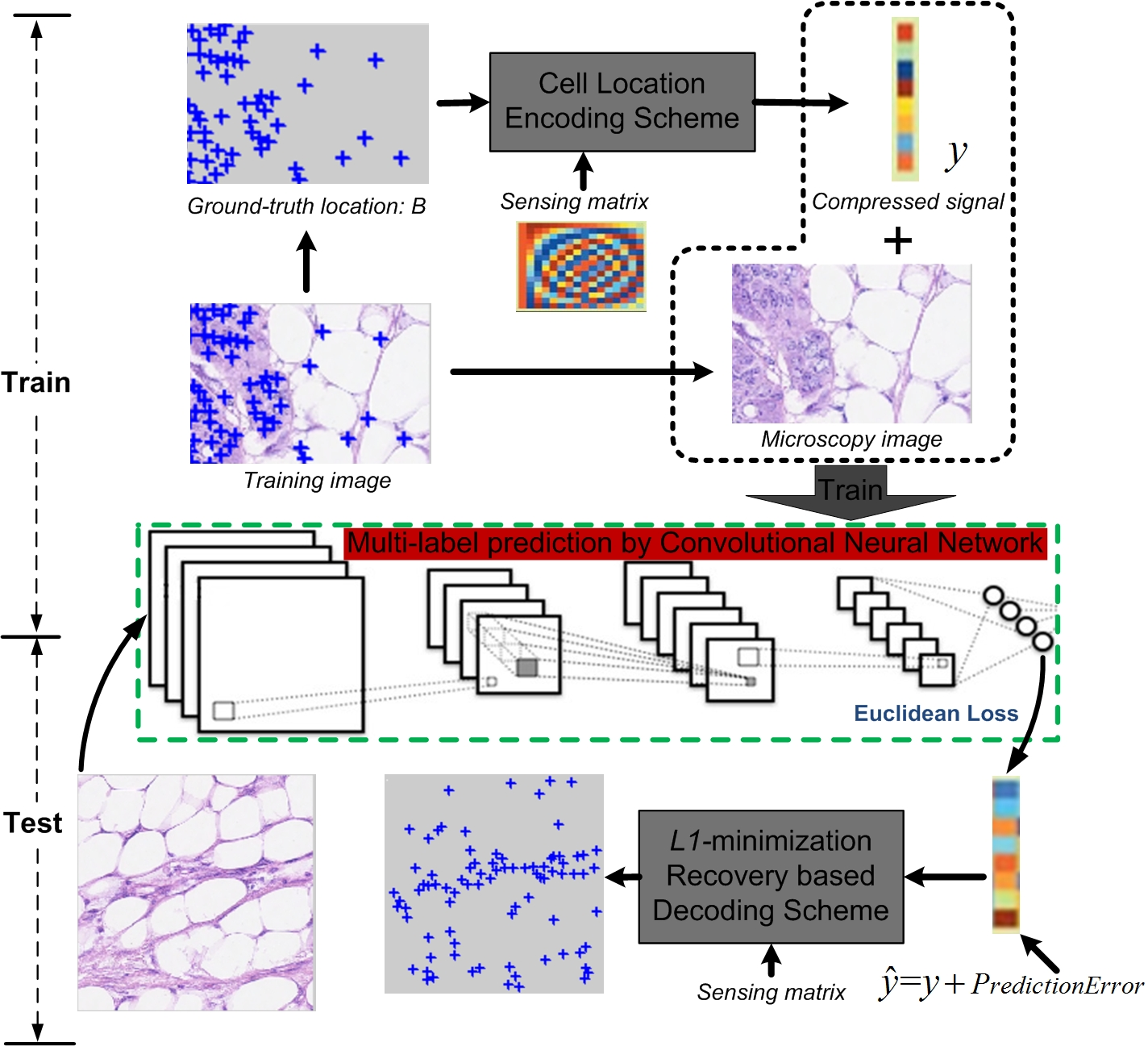
Figure 2: Overview of the CNNCS framework for cell detection and localization.
Encoding and Decoding
Scheme-1 concatenates the binary annotation map into a sparse vector, which is then projected using a random Gaussian matrix. Scheme-2 projects cell centroids onto multiple uniformly distributed axes, encoding their signed distances and concatenating the results. The latter reduces the dimensionality of the sensing matrix and introduces redundancy, facilitating robust recovery. Decoding is performed via L1-norm convex optimization, followed by mean shift clustering to aggregate candidate detections and suppress noise.
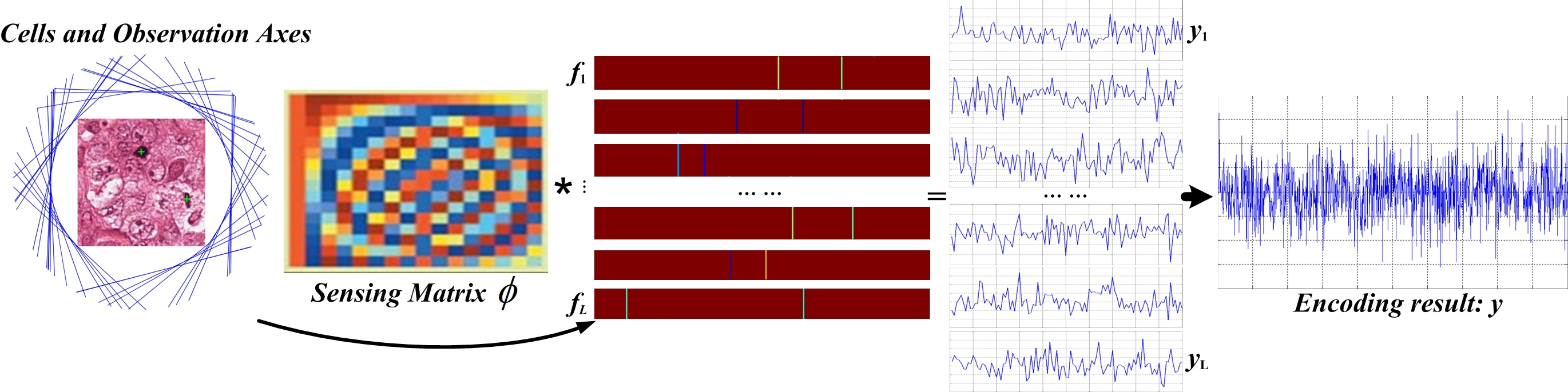
Figure 3: Cell location encoding by signed distances (Scheme-2).
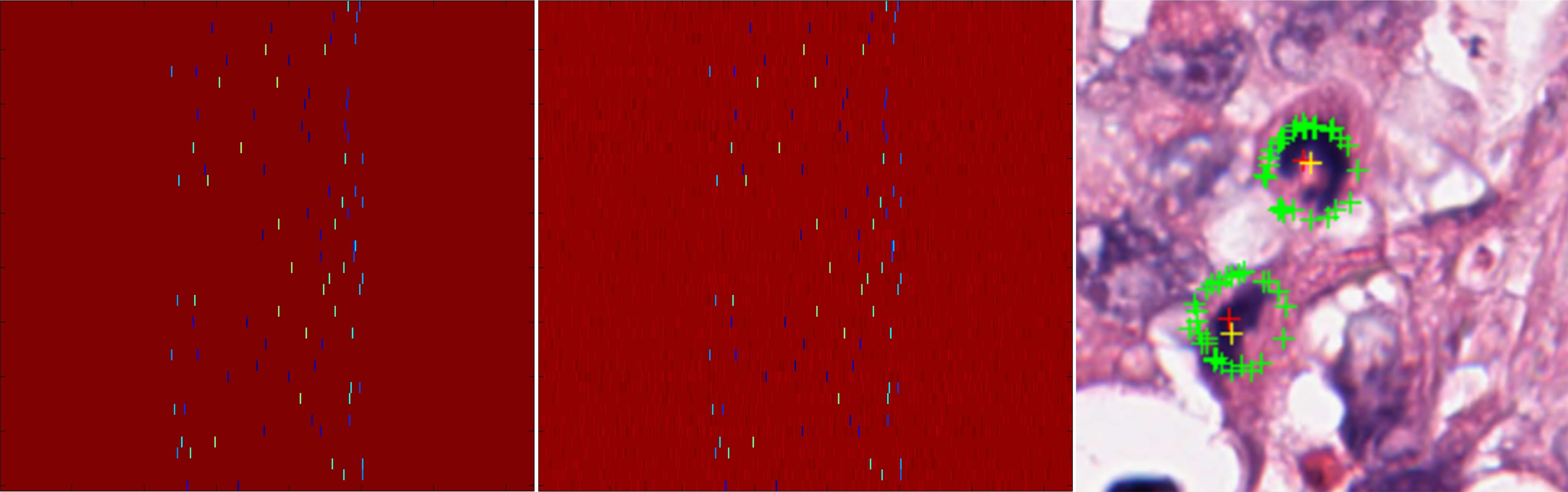
Figure 4: Decoding scheme illustrating true and recovered location signals and final detection results.
CNN Regression and Multi-Task Learning
The regression model utilizes AlexNet and ResNet architectures, with the output layer dimension matching the compressed signal length. Training employs Euclidean loss, and data augmentation via patch rotation enhances robustness. Multi-task learning (MTL) is incorporated by fusing cell count information with the compressed signal, further optimizing detection and counting performance.
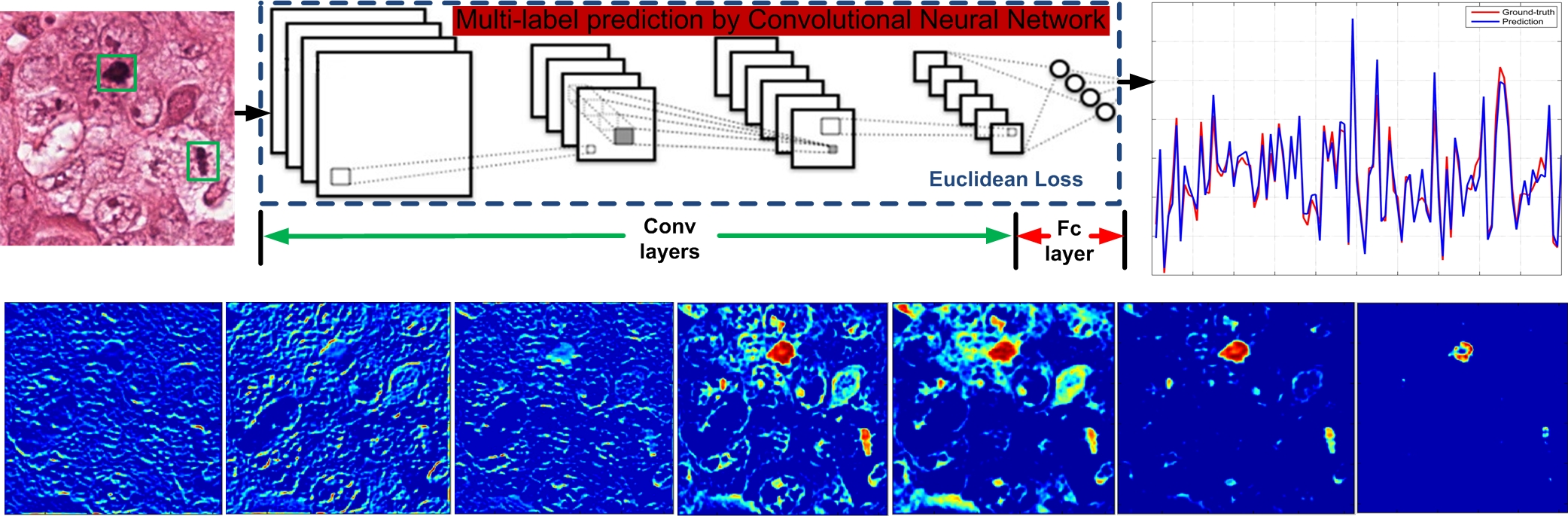
Figure 5: Signal prediction process and feature maps from CNN layers, showing close approximation of ground-truth and predicted compressed signals.
Theoretical Analysis
The equivalence of optimization targets in compressed and original pixel spaces is established via the RIP of the sensing matrix. The generalization error bound demonstrates that the expected error in cell localization is controlled by the predictor's error in compressed space and the reconstruction algorithm's error, justifying the use of deep CNNs and advanced L1 recovery methods.
Experimental Results
Datasets and Evaluation
Seven datasets, including ICPR 2012/2014, AMIDA 2013/2016, and others, are used for evaluation. The annotation format and dataset diversity are illustrated below.
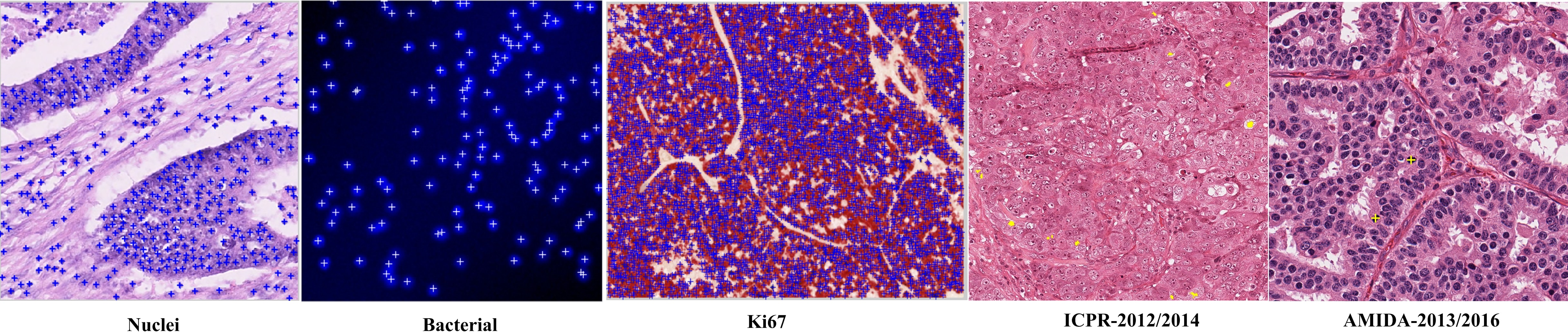
Figure 6: Examples of datasets and their cell centroid annotations.
Performance is measured using precision, recall, and F1-score, with detection considered correct if the predicted centroid lies within a specified radius of the ground truth.
Comparative Analysis
Scheme-1 Results
CNNCS with Scheme-1 outperforms traditional methods (FCN-based, Le.detect, CasNN) in precision-recall curves, particularly at higher recall values, due to its robustness against prediction bias and error in dense cell scenarios.
Scheme-2 Results
On the ICPR 2012 dataset, CNNCS-ResNet-MTL achieves the highest F1-score (0.837), surpassing previous state-of-the-art methods. The redundancy in Scheme-2 encoding and ensemble averaging contribute to improved precision and recall. On the ICPR 2014 dataset, CNNCS nearly doubles the F1-score of the second-best team (0.633 vs. 0.356). On AMIDA 2013/2016, CNNCS consistently ranks in the top three, demonstrating generalizability across diverse histology images and cell types.
Discussion
The integration of compressed sensing with deep CNNs for output space encoding represents a significant methodological advancement for sparse object detection tasks. The approach mitigates class imbalance and leverages the theoretical guarantees of CS for stable recovery. The redundancy in Scheme-2 encoding and ensemble averaging strategies further enhance detection reliability, particularly in challenging, high-density scenarios. The empirical results indicate that CNNCS is competitive with, and often superior to, existing methods across multiple datasets and contest benchmarks.
The framework's modularity allows for the adoption of advanced CNN architectures and multi-task learning, suggesting potential for further improvements via end-to-end training and more sophisticated ensemble techniques. The theoretical analysis provides a foundation for extending the approach to other sparse detection problems in medical imaging and beyond.
Conclusion
The CNNCS framework demonstrates that deep convolutional neural networks, when combined with compressed sensing-based output encoding, can effectively address the challenges of cell detection and localization in microscopy images. The method achieves top-tier performance on multiple benchmarks, with strong theoretical justification and practical advantages in memory efficiency and robustness. Future work may focus on fully end-to-end training and adaptation to other domains requiring sparse object detection.







